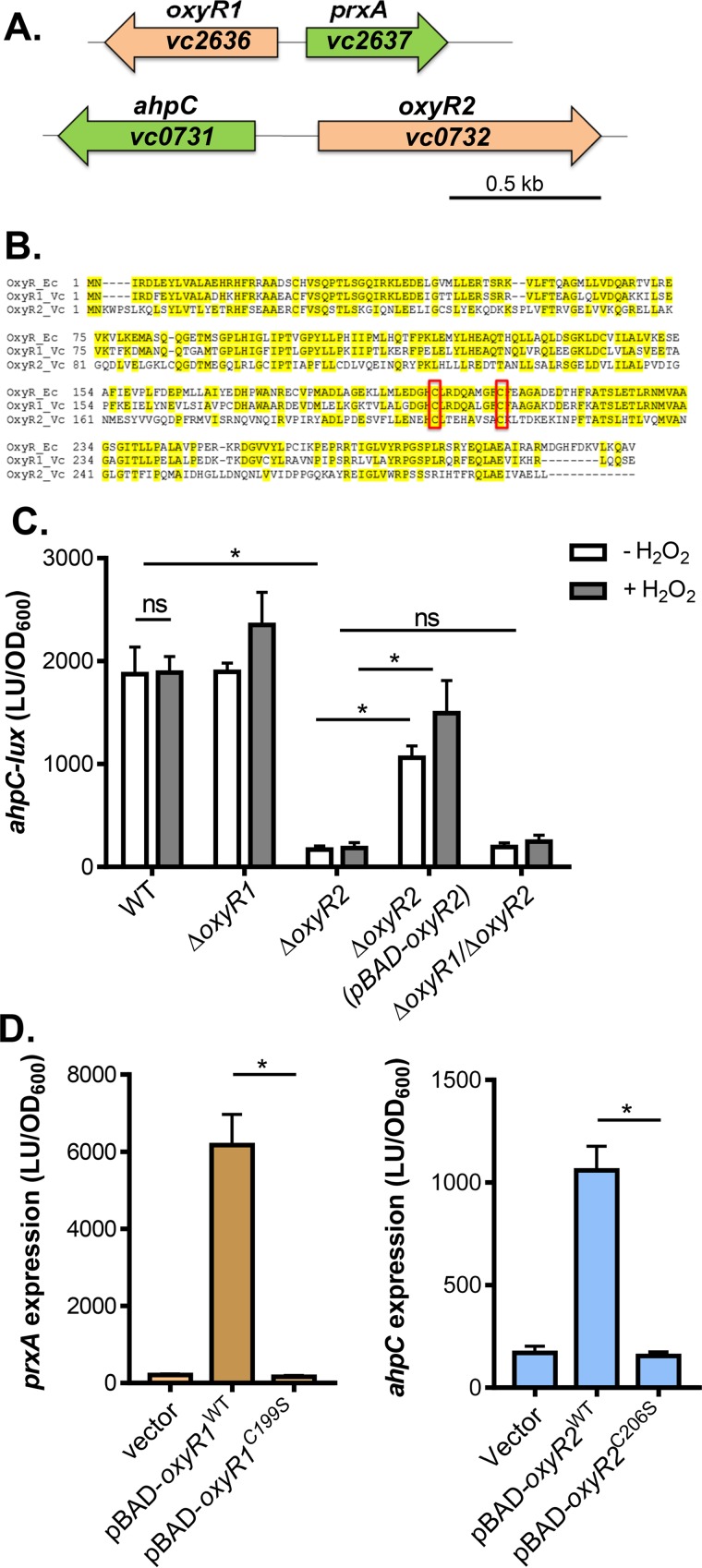
FIG 1.
OxyR2 regulation of ahpC. (A) Schematic representation of the genetic structure of oxyR1-prxA and oxyR2-ahpC regulons. (B) Sequence alignment of E. coli OxyR (OxyR_Ec), V. cholerae VC2636 (OxyR1_Vc), and V. cholerae VC0732 (OxyR2_Vc). Conserved cysteine residues are boxed. (C) ahpC expression. Wild type, ΔoxyR1 ΔoxyR2 mutants, or ΔoxyR1ΔoxyR2 mutants containing the PahpC-luxCDABE transcriptional fusion plasmids were grown standing in AKI medium at 37°C for 4 h followed by shaking for 1 h until mid-log phase was reached. H2O2 (50 μM final concentration) was added to indicated cultures, and cultures were shaken at 37°C for 2 h. Luminescence was then measured and reported normalized to OD600. LU, luminescence unit. For complementation assays, 0.1% arabinose was included in the medium to activate pBAD-oxyR2. (D) The importance of cysteine residues in OxyR1 (left) and OxyR2 (right). pBAD-oxyR1WT or pBAD-oxyR1C199S was introduced into ΔoxyR1 (pPprxA-luxCDABE), and pBAD-oxyR2WT or pBAD-oxyR2C206S was introduced into ΔoxyR2 (pPahpC-luxCDABE). Arabinose (0.1%) was used to induce the PBAD promoter, and 50 μM H2O2 (final concentration) was added to activate OxyR1. Data shown represent the mean and standard deviation from three biological replicates. ns, not significant; *, Student t test P value of <0.05.