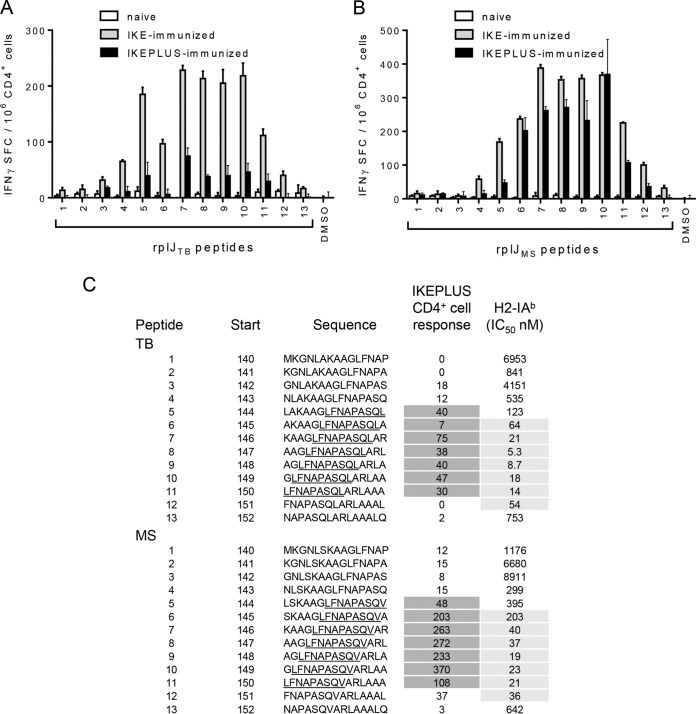
FIG 3.
Mapping of the core epitope of RplJ. C57BL/6 mice were immunized with 5 ×107 CFU IKE or IKEPLUS i.v. or 5 ×106 CFU BCG-Danish s.c. Two weeks later, CD4+ T cells were isolated from the spleen and assayed by an IFN-γ ELISPOT assay for responses to overlapping RplJ 15-mer peptides designed around the original RplJ epitope (peptide 7) and representing either M. tuberculosis H37Rv (TB) or M. smegmatis mc2155 (MS) sequences. (A) IFN-γ ELISPOT responses to M. tuberculosis peptides by CD4+ T cells from IKE-immunized, IKEPLUS-immunized, or naive mice. DMSO was used as a negative control. (B) Same as for panel A except showing responses to M. smegmatis peptides. (C) Sequences of both M. tuberculosis and M. smegmatis peptides assayed in panels A and B are shown, along with the location of their first residue (start) in the RplJ protein. The sequences are shown in single-letter amino acid code. With respect to CD4+ T cell responses, the minimal core epitopes for the M. tuberculosis and M. smegmatis sequences were defined as the common residues among the reactive peptides (underlined). Responses of immune CD4+ T cells to the peptides are summarized as the number of positive cells per 106 CD4+ T cells, quantified by an IFN-γ ELISPOT assay, as shown in panels A and B, and values for reactive peptides are indicated by dark shading. Binding affinities of the peptides for I-Ab were also determined, and the IC50s for each peptide (in nanomolar) are listed, with values indicative of significant binding being highlighted by gray shading. Data shown in panels A and B represent mean values and standard errors for triplicate samples. All data shown are representative of results from two independent experiments.