Figure 1.
Hierarchy of TPA-Induced Histone Modifications at TSS Regions in MEFs
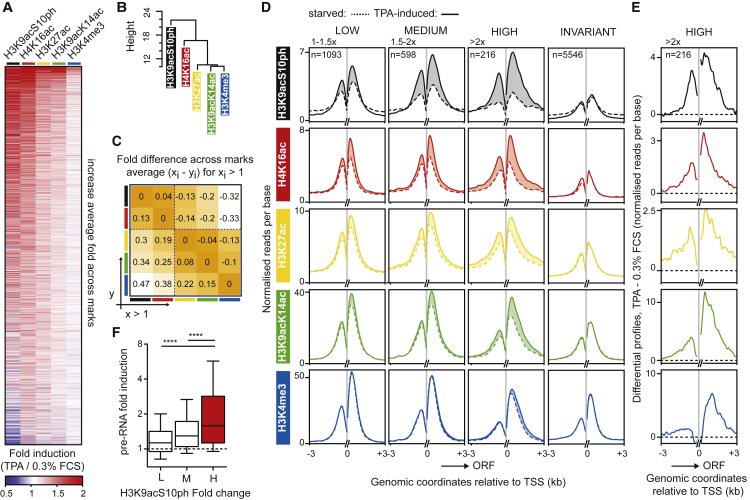
Histone modifications at annotated TSSs were assessed by ChIP-seq using antibodies H3K9acS10ph, H4K16ac; H3K27ac, H3K9acK14ac, and H3K4me3.
(A) The heatmap shows changes in ChIP-seq signal across at 2,364 TSS regions (−2 to +1 kb), where at least one modification shows a statistically significant change according to DESeq (among ∼12,000 with detectable ChIP-seq signal). Ranking is by average fold change.
(B) Dendrogram plot for the five histone modifications (distance method “euclidean,” nesting method “single”).
(C) Average difference in fold induction per compared pair of histone modifications (average of xi − yi, for xi > 1).
(D) Metaprofiles for the 2,364 TSS regions, grouped by change in H3K9acS10ph (low, 1–1.5× [n = 1,093]; medium, 1.5–2× [n = 598]; high, ≥2× [n = 216]) and TSS regions exhibiting unchanged histone modifications (≥100 reads across each TSS; n = 5,546). ChIP-seq read counts extend 3 kb either side of the TSS. 150 bp on each side of the TSS, where low read counts presumably reflect nucleosome depletion, were excluded. Shaded areas denote difference between the TPA-induced and uninduced levels.
(E) Differential profiles (TPA −0.3% FCS) per base at TSS regions displaying >2× increased H3K9acS10ph. For other TSS region classes, see Figure S2C.
(F) Boxplot representation of average fold induction of precursor RNA (intronic RNA-seq reads) of the TSS-associated genes as a function of H3K9acS10ph fold change (middle line, median; top and bottom edges, 75th and 25th percentiles).