Figure 2.
Most TPA-Induced Histone Modifications Are TCF Dependent
Histone modifications at annotated TSSs were assessed in wild-type and TKO MEFs as in Figure 1.
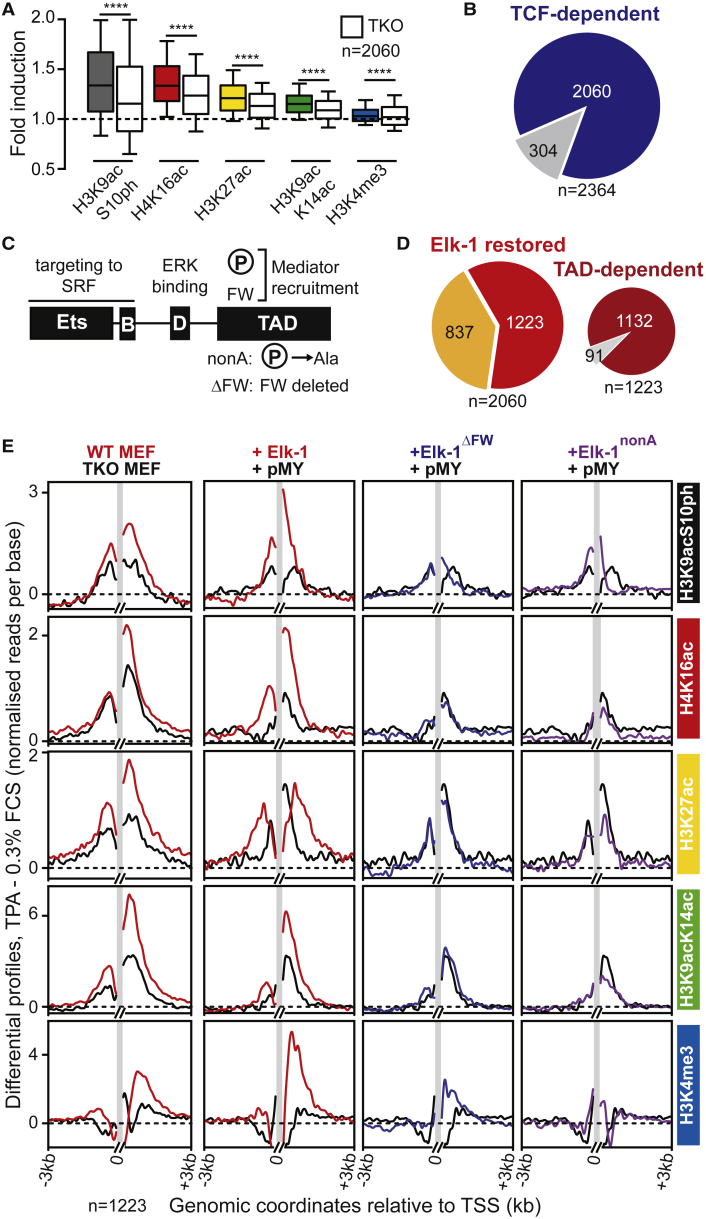
(A) Comparison of ChIP-seq signal in wild-type and TKO MEFs (colored and white boxes, respectively) at the 2,060 TCF-dependent TSS regions. Middle line, median; top and bottom edges, 75th and 25th percentiles; horizontal bars, 90th and 10th percentiles. Statistical significance by Wilcoxon matched-pairs signed rank test (∗∗∗∗p < 0.0001).
(B) TCF dependence of TPA-induced changes in histone modifications.
(C) Domain structure of Elk-1 (Buchwalter et al., 2004): in Elk-1nonA, all ERK sites in the activation domain (TAD) are substituted by alanine; Elk-1ΔFW lacks the FW motif required for Mediator recruitment (Balamotis et al., 2009).
(D) Proportion of TSS regions where expression of wild-type Elk-1 restored regulated histone modifications in TKO MEFs.
(E) Differential profiles, as in Figure 1E, of ChIP-seq signals across the 1,248 TSS regions where regulated histone modifications is restored by Elk-1. Left: profiles in wild-type (red) and TKO MEFs (black); right: differential profiles in TKO MEFs reconstituted with wild-type Elk-1 (red), Elk-1ΔFW (blue), Elk-1nonA (purple), or pMY vector (black) (Gualdrini et al., 2016).
See Figure S4 for full data.