Figure 6.
siRNA Screen Defines an Ordered Series of TCF-Dependent Chromatin Steps Required for Egr1 and Fos Activation
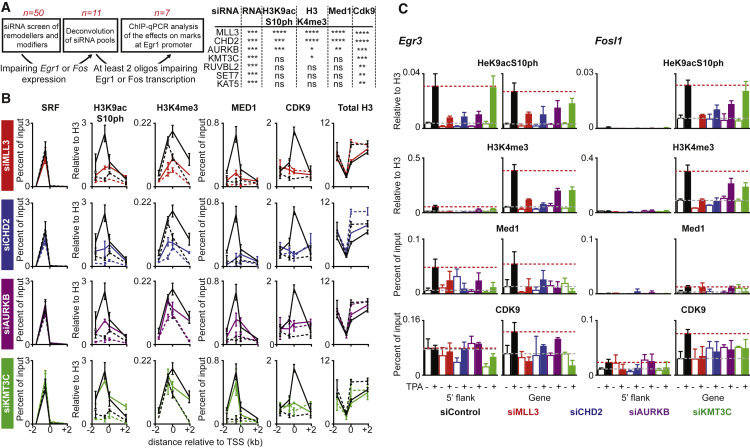
(A) Left: siRNA screening strategy, with number of candidate hits remaining at each stage indicated. Right: summary of siRNA effects on transcription or ChIP signals at the 5′-flanking or TSS-proximal region of Egr1 (one-way ANOVA, corrected for multiple comparisons: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; ns, not significant).
(B) Quantitative ChIP at Egr1 with the indicated antibodies following depletion of MLL3 (red), CHD2 (blue), AURKB (purple), and KMT3C (green) or scrambled oligonucleotide control (black). Histone ChIP signals were normalized to H3.
(C) Quantitative ChIP at Egr1, Egr3, and Fosl1 analyzed using 5′-flanking or TSS-proximal PCR probes (see Figure 4A) following depletion of MLL3 (red), CHD2 (blue), AURKB (purple), and KMT3C (green) or scrambled oligonucleotide control (black). Histone ChIP signals were normalized to H3. Data are mean ± SEM; n = 3.