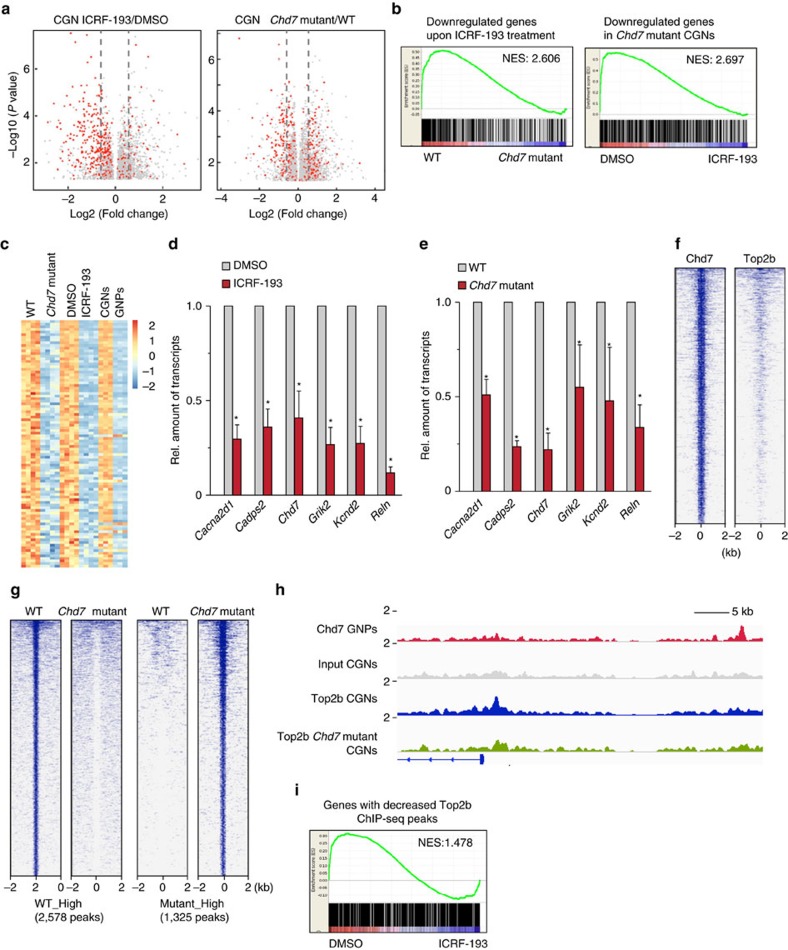
Figure 6. Chd7 recruits Top2b to activate long neuronal genes.
(a) Volcano plots depict gene expression changes (P<0.05) between CGNs treated with ICRF-193 or DMSO (left panel) and between WT [Chd7f/f] and Chd7 homozygous mutant [Atoh1-Cre::Chd7f/f] CGNs (right panel). Genes longer than 100 kb are labelled as red dots. The grey lines show the position of fold change of 1.5. (b) Gene Set Enrichment Analysis (GSEA) of genes that were significantly downregulated (P<0.01) in CGNs upon ICRF-193 treatment in WT and Chd7 mutant CGNs (left panel) and were significantly downregulated in Chd7 mutant CGNs (P<0.01) in ICRF-193- and DMSO-treated WT CGNs (right panel). P<0.001. NES: Normalized Enrichment Score. (c) Heatmap of the expression of genes activated by both Chd7 and Top2b in WT and Chd7 mutant CGNs, and ICRF-193- or DMSO-treated CGNs and cells cultured with (GNPs) or without SAG (CGNs). The colour scale is shown on the right. (d) qRT–PCR analysis represented Top2b-activated genes in CGNs. The level of Gapdh transcripts was used for normalization. Bars represent normalized mean value ± s.d. from four samples for each group. P=0.0003; 0.001; 0.0035; 0.005; 0.0006; 1.0E-5 (from left to right). (e) qRT–PCR analysis of the represented genes in Chd7 WT and mutant CGNs. The level of Gapdh transcripts was used for normalization. Bars represent normalized mean value ± s.d. from four samples for each group. P=0.001; 1.8E-5; 0.0004; 0.028; 0.035; 0.0016 (from left to right). (f) ChIP-seq density heatmaps for Chd7 in GNPs and Top2b in CGNs within ±2 kb of the Chd7 peak centre. (g) ChIP-seq density heatmaps depict altered Top2b ChIP-seq peaks (P<0.01, fold change>2) between WT and Chd7 mutant CGNs. Regions within ±2 kb of the Top2b peak centre are shown. (h) Track view of ChIP-seq density profile for Chd7 in GNPs and Top2b in WT and Chd7 mutant CGNs in the region upstream of Cadps2 gene. (i) GSEA of selected genes that have at least one decreased Top2b ChIP-seq peak in Chd7-deficient CGNs in ICRF-193- and DMSO-treated WT CGNs. P<0.001.