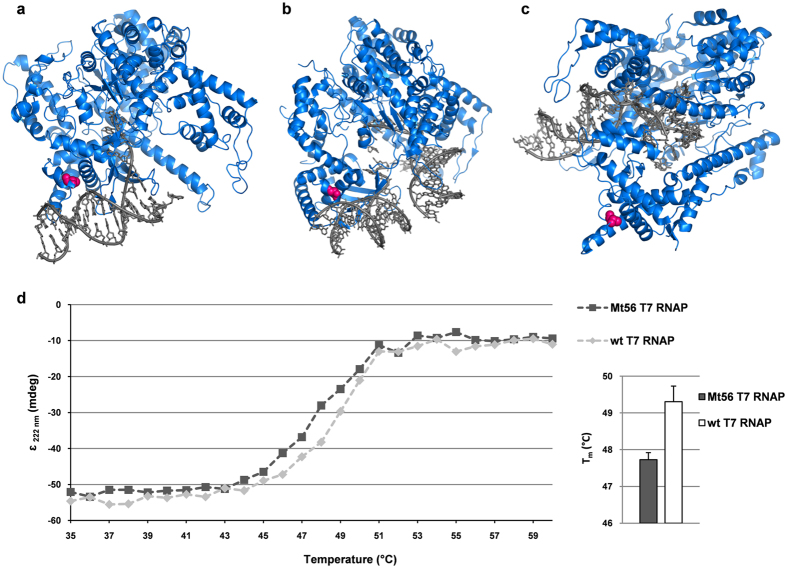
Figure 6. A102D is part of the DNA binding domain of the T7 RNAP and has only a marginal effect on its stability.
To assess the potential effect of A102D on the functioning of the T7 RNAP, available structures of the T7 RNAP in complex with DNA were analyzed. The position 102 in the T7 RNAP structure is highlighted using pink spheres. To monitor the effect of A102D on T7 RNAP stability circular dichroism (CD) melting curves were recorded. (a–c) From the left to the right; structures representing the T7 RNAP bound to the T7 promoter (PDB 1CEZ), the T7 RNAP in transition from initiation to elongation (PDB 3E2E) and the T7 RNAP elongation complex (PDB 1H38)28,29,30. The structures show that position 102 is part the DNA binding site of the T7 RNAP that is involved in the formation of the initial T7 RNAP-T7 promoter complex, but far away from the active site in the elongation complex. (d) Left panel. To monitor the stability of the Mt56 T7 RNAP and wild-type T7 RNAP circular dichroism (CD) melting curves were recorded. CD spectra of the purified wild-type T7 RNAP and Mt56 T7 RNAP were recorded at 222 nm at increasing temperatures. Representative examples are shown. Right panel. Melting temperatures of Mt56 T7 RNAP and wild-type T7 RNAP are based on the CD signal where half the protein was folded. Calculations are based on 3 independent measurements.