Figure 2.
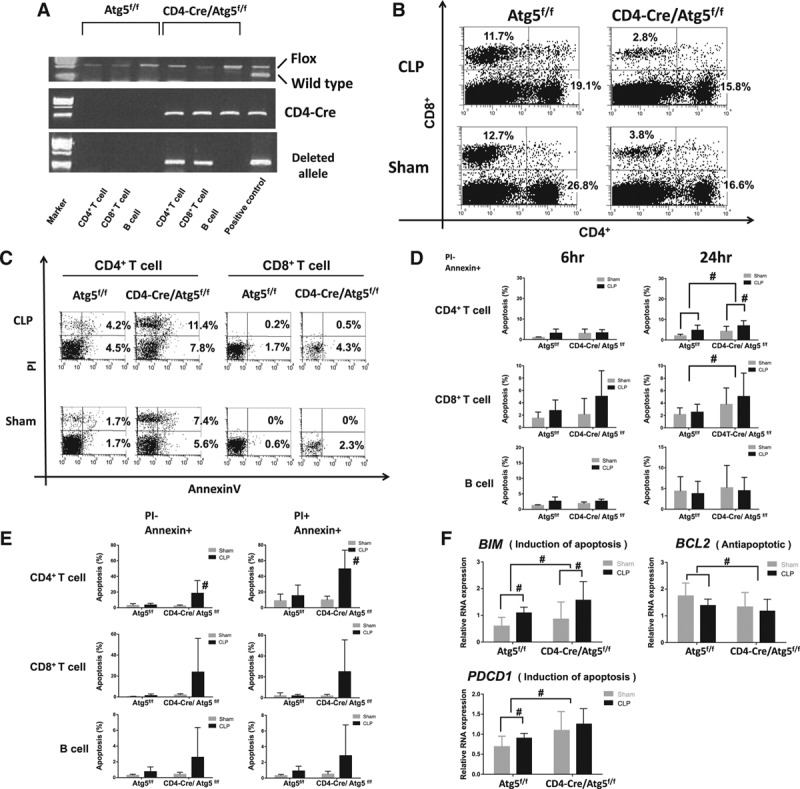
Apoptosis was accelerated by Atg5 deletion in T cells during sepsis. A, Polymerase chain reaction amplification of genomic DNA from sorted lymphocytes in Atg5f/f and CD4-Cre/Atg5f/f mice. Atg5 flox allele and CD4-Cre allele are shown in the upper and middle figure, respectively. The positive controls are tail DNA from CD4-Cre/Atg5f/f mice. In the lower figure, the deleted allele is found in CD4+ T cells and CD8+ T cells extracted from CD4-Cre/Atg5f/f mice. The positive control is liver DNA from liver specific-Cre/Atg5f/f mice. B, Representative subpopulation of splenocytes in Atg5f/f and CD4-Cre/Atg5f/f mice. Mice were performed cecal ligation and puncture (CLP) or sham procedure. Splenocytes were stained with anti-CD4+/PE and anti-CD8+/APC. Representative data of five independent experiments are shown. C, FACS profiles of splenocytes stained with Annexin V and PI in Atg5f/f and CD4-Cre/Atg5f/f mice for 24 hr postoperatively. Samples were stained with anti-CD4+/PE and anti-CD8+/APC. Representative data of five independent experiments are shown. D, Subpopulation of Annexin V positive and PI negative staining splenocytes were shown for early (postoperative period, 6 hr) and late (postoperative period, 24 hr) apoptosis. Data are expressed as mean and sd; n = 8–10 mice in each group; #p < 0.05 was significance analyzed by two-way analysis of variance (ANOVA) and student t test. E, Subpopulation of Annexin V and PI staining lymphocytes from peripheral blood were shown for early (Annexin V positive and PI negative) and late (Annexin V positive and PI positive) phase of apoptosis. Data are expressed as mean and sd; n = 6–8 mice in each group; #p < 0.05 was significance analyzed by student t test between CLP-operated Atg5f/f mice and CLP-operated CD4-Cre/Atg5f/f mice. F, Relative RNA expression for apoptotic gene in sham, CLP-operated Atg5f/f mice and sham, and CLP-operated CD4-Cre/Atg5f/f mice. Total RNA in CD4+ splenic lymphocytes was extracted from experimental mice at 24 hr after the procedure, and then relative RNA expression for the several genes was analyzed. Data are expressed as mean and sd; n = 8–10 mice in each group; #p < 0.05 was significance analyzed by two-way ANOVA and student t test. APC = allophycocyanin, Bcl-2 = B-cell leukemia/lymphoma 2, BIM = Bcl-2-like 11, FACS = fluorescence activated cell sorting, PDCD1 = programmed cell death 1, PE = phycoerythrin, PI = propidium iodide.
