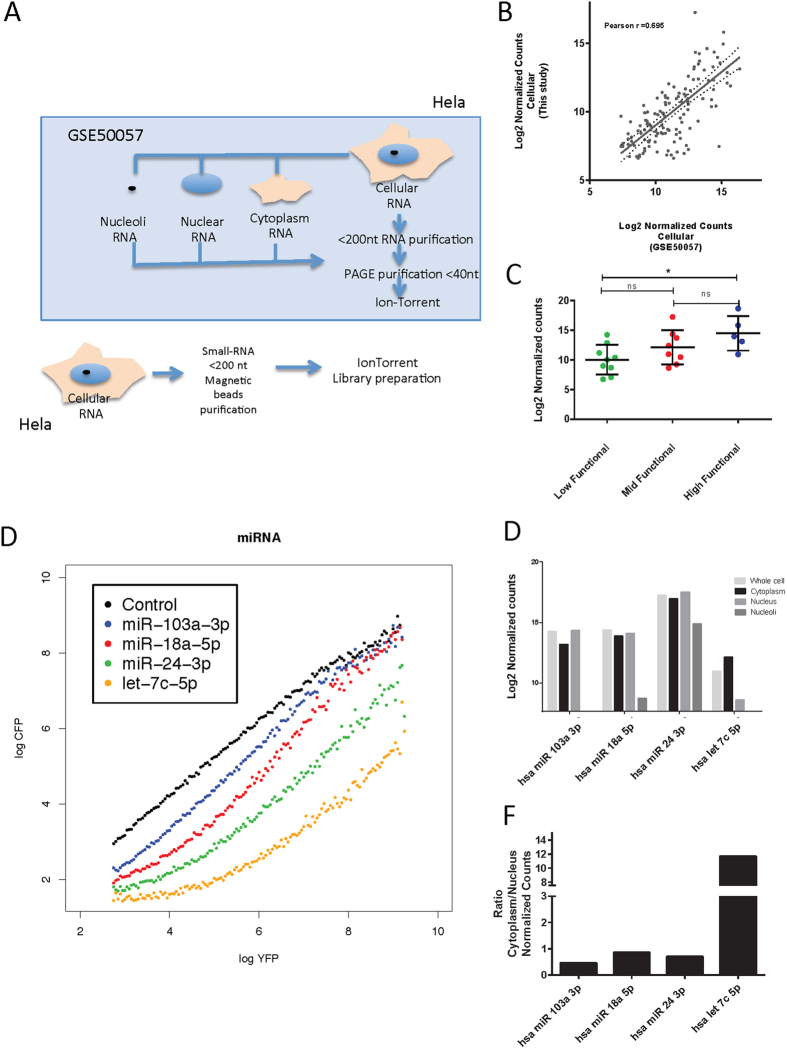
Figure 6. Subcellular localization of miRNAs influences their functional output.
GEO50057 retrieved raw data from HeLa small – RNA seq experiment was retrieved and compared with fresh produced total cell small RNA library. Fractionated cell compartment derived small RNA libraries were used for further analysis of nuclear, cytoplasmic and nucleolar content. (A) Experimental layout for HeLa small RNA library preparation in GSE5005728 and this study. (B) Comparison of small RNA expression profiles from HeLa cells from two independent sources, plot of normalized counts HeLa library prepared vs retrieved (r = 0.695, P < 2.2e16 r2 = 0.48).(C) miRNAs UTA reporters were transfected into HeLa cells, evaluated after 72 h and UTA transfer functions were plotted (Supplementary Fig. S8) distributed into the three ordinal variables as before and cellular normalized counts were compared between groups (P < 0.01 Kruskal-Wallis test, Dunn’s multiple comparisons test * < 0.05). Four individual miRNAs were chosen according to their cellular expression, 3 miRNAs on low and mid functional groups with expression higher than the high functional median (miR-103a-3p, miR-18a-5p, miR-24-3p) and one high functional miRNA lower than the mid functional median of normalized cellular read counts. (D) UTA transfer functions (E) Bar plots for cellular compartments small RNA libraries and (F) Cytoplasmic/Nuclear ratio for selected candidates.