Abstract
Background
In Drosophila early post-meiotic spermatids, mitochondria undergo dramatic shaping into the Nebenkern, a spherical body with complex internal structure that contains two interwrapped giant mitochondrial derivatives. The purpose of this study was to elucidate genetic and molecular mechanisms underlying the shaping of this structure.
Results
The knotted onions (knon) gene encodes an unconventionally large testis-specific paralog of ATP synthase subunit d and is required for internal structure of the Nebenkern as well as its subsequent disassembly and elongation. Knon localizes to spermatid mitochondria and, when exogenously expressed in flight muscle, alters the ratio of ATP synthase complex dimers to monomers. By RNAi knockdown we uncovered mitochondrial shaping roles for other testis-expressed ATP synthase subunits.
Conclusions
We demonstrate the first known instance of a tissue-specific ATP synthase subunit affecting tissue-specific mitochondrial morphogenesis. Since ATP synthase dimerization is known to affect the degree of inner mitochondrial membrane curvature in other systems, the effect of Knon and other testis-specific paralogs of ATP synthase subunits may be to mediate differential membrane curvature within the Nebenkern.
Keywords: Drosophila melanogaster, Spermatogenesis, Mitochondria, ATP synthase, Cristae
Background
Mitochondrial dynamics, including fusion, division, regulated movement, quality control, and interaction with the endoplasmic reticulum, have been characterized largely in yeast and cultured mammalian cells (reviewed in [1, 2]). Studies in specialized cell types with high energy needs in other organisms, such as neurons and sperm in Drosophila melanogaster, also provided early and ongoing insight into molecular mechanisms of mitochondrial shaping [3, 4]. Drosophila spermatogenesis involves a concerted developmentally-controlled program of specialized mitochondrial morphogenesis and thus is an ideal system for exploring molecular underpinnings. Mitochondria aggregate during and after male meiosis, subsequently fusing dramatically into two giant mitochondrial derivatives that interdigitate in layers at the early round spermatid stage to generate a large, onion-like, layered spherical structure called the Nebenkern [5–7]. During subsequent spermatid elongation stages, the mitochondrial derivatives within the Nebenkern unfurl and elongate beside the growing flagellar axoneme. Testis-enriched paralogs of more broadly-expressed genes [8] are common and are often identified via genetic analysis as associated with these functions, since null mutations in the testis-enriched paralogs can result in male-sterile but adult-viable mutant phenotypes.
ATP synthase is a large multi-protein complex, found in prokaryotes as well as in mitochondria and chloroplasts of eukaryotes, that catalyzes ATP formation from ADP and inorganic phosphate during cellular respiration and photosynthesis (reviewed in [9]). In mitochondria, the ATP synthase complex typically contains at least 16 subunits divided among the F1, F0, and peripheral stalk regions. The F1 portion extends into the mitochondrial matrix, physically attaching in two ways to the F0 portion, which is embedded in the inner membrane: via the F1 central stalk that allows rotation, and via the peripheral or stator stalk, which stabilizes the complex. The F0 portion rotates in response to proton movement down the gradient established by the electron transport chain, and the resulting torque and conformational changes in the F1 portion lead to ATP formation. Defects in the ATP synthase complex in humans can lead to neuromuscular disorders (reviewed in [10]) due to deficiencies in ATP production and overall mitochondrial dysfunction.
As shown mainly in yeast and cultured mammalian cells, the positive membrane curvature at the tips of cristae in the inner mitochondrial membrane is mediated in part by higher-order dimerization and oligomerization of ATP synthase complexes (reviewed in [11–14]). These interactions are separable from the metabolic role of ATP synthase, as some mutants with intact single ATP synthase complexes that cannot dimerize have aberrant cristae structure but can still respire [15, 16]. Cryo-electron microscopy indicates that bovine and yeast ATP synthase complex monomers induce 43° membrane curvature, and that the predicted dimer-induced membrane curvature of 86° matches observed values well [17, 18]. Tomography studies indeed show dimer ribbons corresponding with tight membrane curvature [19].
A logical question is whether regulation of ATP synthase higher-order structures helps to mediate variability in inner mitochondrial membrane conformation across cell types and different conditions. In Drosophila spermatogenesis, the large mitochondrial derivatives of the Nebenkern in early round spermatids do not have prominent cristae or sharp positive curvature of the inner mitochondrial membrane; instead the inner and outer mitochondrial membranes appear closely apposed across most of their area and parallel to each other in broad concentric circles of membranes reminiscent of an onion slice (hence early round spermatids are considered to be at the onion stage) [5, 20]. In this way, spermatid wild-type inner mitochondrial membranes resemble yeast inner mitochondrial membranes in cells deficient for ATP synthase dimerization.
Here we report a role for a tissue-specific isoform of ATP synthase subunit d in mediating internal Nebenkern structure within Drosophila melanogaster spermatids. We also provide evidence that other tissue-specific ATP synthase subunit paralogs help shape spermatid mitochondria. Subunit d is a major part of the peripheral stalk that stabilizes the F0 and F1 portions of ATP synthase as they rotate with respect to each other. Subunit d was first described in mammalian mitochondrial ATP synthase [21] and later in yeast [22]. It is not found in bacterial or chloroplast ATP synthase. In S. cerevisiae, subunit d is oriented with its amino terminus farthest from the membrane and the carboxy terminus oriented closer to the inner mitochondrial membrane, spanning the distance between the F0 and F1 portions; while it does not itself span the inner mitochondrial membrane, it interacts via multiple coiled-coil domains with the transmembrane subunit b of the peripheral stalk [23, 24]. We identified an uncharacteristically large ATP synthase subunit d paralog expressed in Drosophila testes that is associated with internal Nebenkern structure and that may act via alteration of ATP synthase dimerization. Our work establishes a connection between tissue-specific variants of ATP synthase subunits and tissue-specific internal mitochondrial structure.
Results
knon mutants show aberrant mitochondrial elongation during spermatogenesis and have faulty Nebenkern internal structure
The knotted onions (knon ms(2)1400) mutant allele of Drosophila melanogaster was identified in a screen for recessive male-sterile mutations via mobilization of a marked P element [25]. Subsequent exposure of the mutant chromosome to transposase yielded no revertants of male sterility out of over thirty independent strains that lost w +-associated eye color, indicating that the spermatogenesis phenotype was likely not associated with an intact P element. Males homozygous for knon ms(2)1400 were viable but completely sterile and had no individualized, motile sperm. Homozygous knon ms(2)1400 females were completely viable and fertile. Phase-contrast microscopy of live testis preparations from knon ms(2)1400 males revealed a failure of the Nebenkern to elongate beside the growing flagellar axoneme (Fig. 1). To distinguish Nebenkern-intrinsic (mitochondrial structure) from Nebenkern-extrinsic (e.g., cytoskeletal or mitochondrial transport defects) sources of elongation failure, we analyzed testes from males double mutant for knon and the dynamin-related GTPase fuzzy onions, fzo, which is required for Nebenkern-associated mitochondrial fusion [3]. We found that knon ms(2)1400 fzo 1 double mutants exhibit the fzo phenotype, wherein many smaller, unfused mitochondria successfully elongate alongside the axoneme, although the elongation appeared to be slightly delayed. This bypass of the elongation defect (Fig. 2, panels a and e) suggests that the knon primary defect may be faulty Nebenkern structure at the onion stage that interferes with the ability of mitochondrial derivatives to unfurl for subsequent elongation. Consistent with this idea, transmission electron microscopy (TEM) of knon ms(2)1400 testes (Fig. 2g) indeed showed aberrant internal Nebenkern structure at the early round spermatid stage (hence the name “knotted onions”). Cross sections of knon ms(2)1400 elongating spermatid cysts showed axonemes without associated mitochondrial derivatives (Fig. 2c). In contrast, elongating knon ms(2)1400 ; fzo 1 spermatids had multiple (though fewer than in fzo alone) mitochondrial derivatives beside elongating axonemes (Fig. 2e). Mitochondria in testes from knon ms(2)1400 males did have a membrane potential as demonstrated by rhodamine 123 staining (Fig. 1h), suggesting that the phenotype appears to be primarily related to defects in mitochondrial shaping rather than in respiration.
Fig. 1.
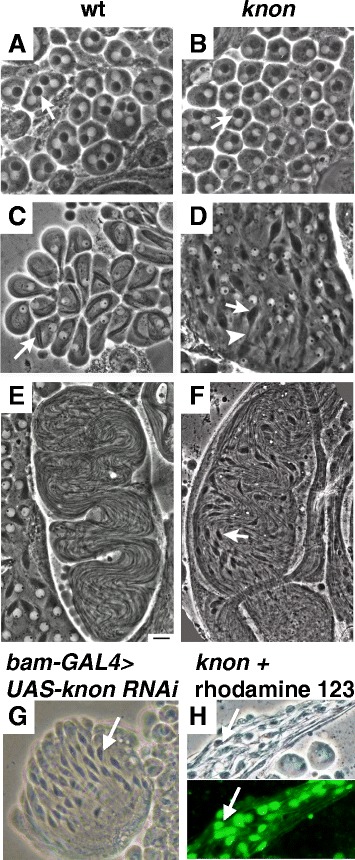
knon mutant and knockdown spermatids show aberrant Nebenkern elongation but have an intact membrane potential. Phase-contrast micrographs of live squashed testis preparations showing post-meiotic spermatids from wild-type (a, c, and e) and homozygous knon ms(2)1400 males (b, d, f, h) at the early round (onion) stage (a-b), the early elongation stage (c-d), and later in spermatid elongation (e-f). g RNAi knockdown of knon in the testis recapitulates the mutant phenotype, with unelongated Nebenkerne (arrow) in nearly mature elongating spermatid cysts. h Paired phase-contrast and fluorescence micrographs of an elongating spermatid cyst from a knon ms(2)1400 /Df(2R)7E male. Rhodamine 123 uptake by the unelongated mitochondrial derivatives (arrow) in a nearly mature spermatid cyst indicates ongoing respiratory activity. Scale bar 20 μm
Fig. 2.
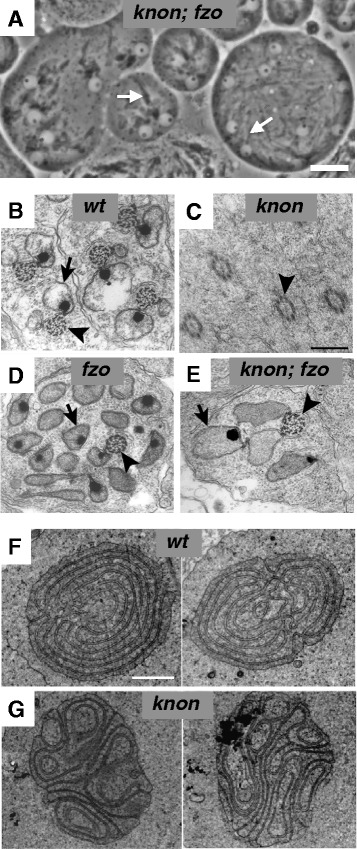
knon mutants exhibit aberrant Nebenkern structure and defective mitochondrial elongation that can be suppressed by bypassing mitochondrial fusion. a Phase-contrast micrograph of early elongation-stage spermatids from knon ms(2)1400 ; fzo 1 males show some degree of elongation of unfused mitochondria (arrows); compare to Fig. 1c-d. b-e Cross sections of elongating spermatids viewed by transmission electron microscopy showing axonemes (arrowheads) and elongating mitochondrial derivatives (arrows). Wild-type spermatids (b) show two mitochondrial derivatives per axoneme. c knon ms(2)1400 spermatids lack detectable mitochondrial derivatives beside the axoneme. d fzo 1 spermatid has multiple mitochondrial derivatives per axoneme resulting from failure of mitochondrial fusion to form the Nebenkern [3]. e Spermatid from knon ms(2)1400 ; fzo 1 male shows several mitochondrial derivatives elongating beside the axoneme. Transmission electron micrographs of cross sections of Nebenkerne from wild-type (f) and knon ms(2)1400 (g) males. Two examples of each are shown. Internal “onion” structure is disrupted in knon ms(2)1400 . Scale bars 20 μm in a, 0.5 μm in b-e, and 2 μm in f-g. b and d adapted from [3]
knon is CG7813
We originally mapped knon by recombination to 2.1 ± 0.7 cM distal of curved (polytene region 52D2-9 on chromosome 2R), based on 38 recombinants between curved and welt. A w +-marked P element on the knon ms(2)1400 chromosome mapped near black, approximately 30 cM away, confirming its lack of association with the spermatogenesis phenotype. Further recombination mapping placed knon 0.67 ± 0.07 cM distal to Khc k13219 and 0.36 ± 0.07 cM proximal to Cdk4 k06503, suggesting polytene interval 53B-C as the region of interest. Df(2R)7E (generated through imprecise excision of veg k03402), which lacks ~134 kb distal of the veg k03402 insertion site, failed to complement knon ms(2)1400, showing a phenotype indistinguishable from knon ms(2)1400 homozygotes. In contrast, Df(2R)BSC550 [26] complemented knon ms(2)1400. Together, the breakpoints of Df(2R)7E and Df(2R)BSC550 defined the knon region as including two genes, Sema2a and CG7813 [27]. Sema2a is expressed in many tissues [28] and known to function in the nervous system [29]. FlyAtlas data indicate that CG7813 is expressed highly in the testis and at near-zero levels in adult carcass (minus the germline) and all other tissues tested except larval fat body, in which the larval gonad is embedded [28]. modENCODE RNAseq data indicate that CG7813 is expressed highly in wandering third instar larvae and adult males but not at all in adult females [30], consistent with expression only in the male reproductive tract. Together these data suggested CG7813 as the best candidate for knon.
Rescue experiments and phenocopy by RNAi knockdown confirmed CG7813 as the gene mutated in knon ms(2)1400. Transgenic male flies homozygous for knon ms(2)1400 and carrying an exogenous copy of CG7813 under its own regulatory regions were fertile and exhibited wild-type mitochondrial shaping and elongation during spermatogenesis (Fig. 3). Knocking down CG7813 in the germline using the bam-GAL4 driver [31] and the P[GD12291]v22566 CG7813 RNAi insertion from the Vienna Drosophila Resource Center [32] resulted in sterile males with large unelongated masses of mitochondria at the base of each elongating post-meiotic spermatid, similar to the knon ms(2)1400 phenotype (Fig. 1). Neither the RNAi transgene alone nor the bam-GAL4 transgene alone had any detectable effect on spermatogenesis or male fertility (not shown). We amplified and sequenced the coding region of CG7813 from knon ms(2)1400 homozygotes and found no difference compared to the reference sequence [27], while an upstream region was not amplifiable in knon ms(2)1400. It is therefore likely that the knon ms(2)1400 lesion is in an upstream regulatory region that was disrupted, possibly by P-element insertion and subsequent excision during the original mutant screen [25].
Fig. 3.
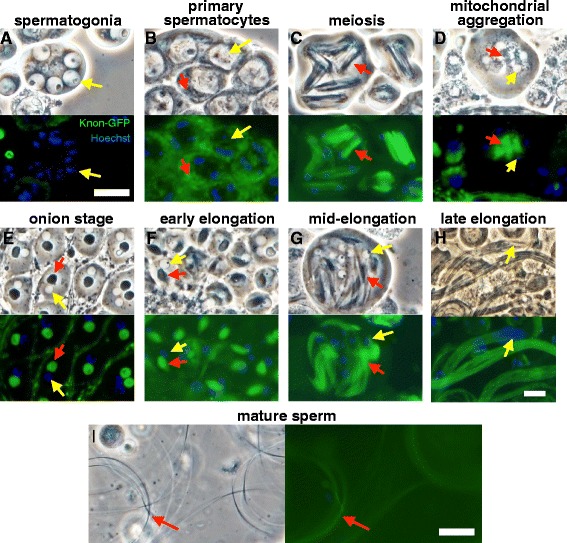
Knon-GFP rescues the mutant phenotype and localizes to mitochondria in primary spermatocytes and post-meiotic spermatids. Phase-contrast and paired fluorescence images of cells from testes of knon ms(2)1400 /Df(2R)7E; knon-GFP/+ males, stained with Hoechst. Knon-GFP (green) was not detectable in spermatogonia (a) but was associated with phase-dark mitochondria (red arrows) in all later stages (b-i). Yellow arrows indicate nuclei; red arrows indicate mitochondria-associated Knon-GFP. In primary spermatocytes (b), Knon-GFP-labeled mitochondria were small and diffuse in the cytoplasm. In meiotic cells (c), Knon-GFP-marked mitochondria associated with the spindle; immediately after meiosis (d) mitochondria aggregated beside each nucleus and were associated with a strong Knon-GFP signal. The mitochondrial derivatives within the Nebenkern at the onion stage (e, red arrow) and during early- (f) and mid-elongation (g) showed unambiguous Knon-GFP localization. In nearly mature elongating spermatid cysts (h), elongated flagella appear wild type with no clumped mitochondrial derivatives (compare to Fig. 1). Knon-GFP is evenly distributed throughout the elongated spermatids. After sperm individualization, motile sperm (i) retain detectable Knon-GFP (red arrow). The apparent syncytial nature of some cells at earlier stages is an artefact of the preparation—ring canals connecting cells in a cyst are commonly burst open by the pressure from a cover slip. These localization results are identical to those seen when Knon-GFP is expressed in a wild-type background (not shown). Scale bars in a-g, h, and i are 20 μm
The MiMIC (Minos Mediated Integration Cassette) insertion line Mi[MIC]CG7813 Mi10492 [33, 34] is a hypomorphic allele of knon (CG7813) and is henceforth referred to as knon Mi10492. The knon Mi10492 insertion is in the coding region of CG7813 between codons 554 and 555 [27] and is therefore predicted to truncate the last 180 amino acids of Knon. Since the element is in reverse orientation with respect to the knon coding sequence, it does not provide a GFP tag to the truncated protein. Homozygous knon Mi10492 males were fertile but at slightly reduced levels, giving rise to 85% as many progeny as wild-type males (n = 56; P = 1.29x10-6), and show incompletely penetrant mild mitochondrial elongation defects (not shown). Males with knon Mi10492 in trans to either Df(2R)7E or knon ms(2)1400 were completely sterile with a mitochondrial elongation defect in spermatids similar to that in knon ms(2)1400 homozygotes, further confirming that the original knon ms(2)1400 phenotype is due to a lesion in CG7813.
Knon is a testis-enriched ATPsynD paralog with large C-terminal extension and associates with spermatid mitochondria
The knon gene encodes a 734 amino acid protein with an amino terminal region 32% identical and 50% similar to Drosophila ATPsynD (178 amino acids), the broadly-expressed canonical d subunit of the ATP synthase complex. Subunit d is part of the peripheral (or stator) stalk connecting the F0 and F1 portions of the complex. Knon, a testis-enriched paralog of subunit d, has 532 additional amino acids at its carboxy terminus beyond the region of homology. BLASTp and tBLASTn searches revealed that this C-terminal region was not homologous to other characterized gene products [35]. Curiously, in this C-terminal region 28% of the residues are either glutamic acid or lysine, making it highly charged, and there are no predicted transmembrane domains or other motifs. The IUPred tool [36] predicts that most of this region is intrinsically unstructured.
Knon-GFP expressed under the control of endogenous knon regulatory sequences rescued the mutant phenotype (confirming functionality and relevance of localization) and was associated with mitochondria during later stages of spermatogenesis (Fig. 3). Knon-GFP was not detectable in the apical testis, which contains the stem cell populations and mitotically proliferating spermatogonia. The protein was detectable starting in spermatocytes during chromosome condensation. The timing of transition from absence of Knon-GFP to abundance was consistent with established patterns of transcription in primary spermatocytes [37]. Knon-GFP co-localized with phase-dark mitochondria throughout spermatogenesis, associating with meiotic spindles, post-meiotic aggregating mitochondria, and Nebenkerne in the onion stage. Knon-GFP persisted on mitochondrial derivatives during and after mitochondrial elongation in elongated spermatid cysts. Mature, individualized wild-type sperm also contained the protein.
Exogenous expression of Knon in flight muscle alters the ratio of ATP synthase complex dimers to monomers but does not affect gross mitochondrial shaping or function
Because the ATP synthase d subunit is located at the interface of bound ATP synthase complexes in a dimer, we hypothesized that the extended C-terminal domain of Knon might favor the monomeric ATP synthase configuration by a steric mechanism. To test whether incorporation of Knon into ATP synthase inhibits dimerization of the complex, we exogenously expressed full-length Knon in flight muscle with the GAL4-UAS system [38] and analyzed ATP synthase complexes by blue native polyacrylamide gel electrophoresis (BN-PAGE) and immunoblotting. (Testis tissue is not amenable to this approach since each dissected testis includes cells at multiple stages, and since the dramatically varying mitochondrial conformations in spermatids make isolation of the organelle impractical.) We generated flies carrying the wild-type knon coding region, with or without a C-terminal GFP tag, downstream of the UAS regulatory region, and crossed these to flies with the flight muscle Act88F-GAL4 driver. We confirmed by fluorescence microscopy robust Act88F-GAL4-induced Knon-GFP expression in flight muscle (not shown). By BN-PAGE and immunoblotting using an antibody specific for ATP synthase subunit α, we examined the internal ratio of ATP synthase dimers to monomers in flies with Act88F-GAL4 alone compared to Act88F-GAL4 driving Knon expression from each of two independently-inserted UAS-knon transgenes (Fig. 4). Quantification of band intensity indicated a statistically significant (P < 0.05) shift toward monomers when Knon is expressed, compared to a control. Such a shift was not seen when we used the Act88F-GAL4 driver to drive RNAi constructs knocking down the broadly-expressed ATPsynD or ATPsynG, though these conditions led to a more pronounced F1 band and a smear at lower molecular weights suggesting increased dissociation of the complex into F0 and F1 portions and subsequent degradation. To test whether exogenous Knon would exert a stronger effect if less of the paralogous ATPsynD was present, we simultaneously expressed Knon and knocked down ATPsynD; the shift toward ATP synthase monomers was similar to expressing Knon alone (Fig. 4a), although with the more pronounced lower molecular weight smear as seen in the knockdown alone. In each case, when Knon was expressed in flight muscle, the BN-PAGE immunoblot also showed bands slightly heavier than the typical monomer bands (Fig. 4a, arrow), perhaps indicating monomers of the ATP synthase complex that incorporated Knon (with its extra 532 amino acids) instead of ATPsynD. While native gels do not allow for precise calculation of molecular mass from distance traveled, the position of the extra band could plausibly represent monomers with an extra 62 kD.
Fig. 4.
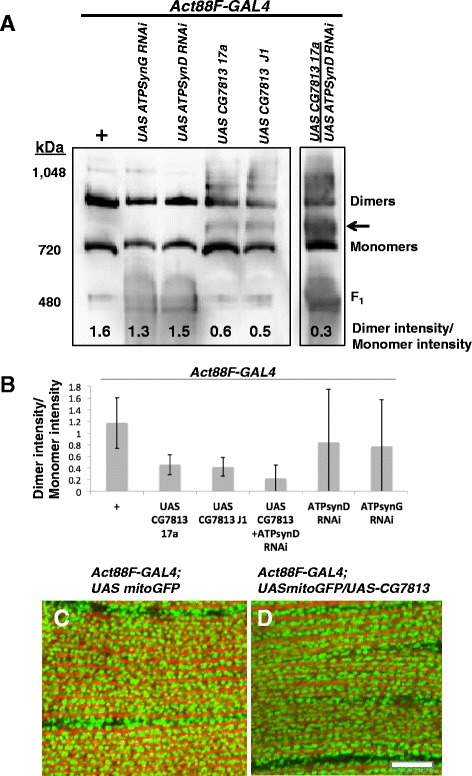
Knon expression in flight muscle alters ATP synthase dimer:monomer ratio but not gross mitochondrial morphology. a Immunoblot using anti-ATP synthase α subunit antibodies on a BN-PAGE gel of mitochondrial isolates from flight muscle of the indicated genotypes. The Act88F-GAL4 flight muscle driver was used as a control (first lane), as well as to drive exogenous Knon (CG7813) expression from two independent transgene insertions (fourth and fifth lanes), RNAi knockdown constructs of ATP synthase subunits g (second lane) and d (third lane), or simultaneous exogenous CG7813 expression and knockdown of ATP synthase subunit d (sixth lane). Dimer/monomer intensity ratios are indicated at bottom. Expressing Knon (CG7813), either with or without concurrent knockdown of the broadly-expressed ATPsynD, reduced the amount of ATP synthase dimers relative to monomers and gave rise to detectable bands of slightly larger size (arrow) than the typical monomer bands. b Quantification of dimer/monomer band intensity ratios from multiple biological replicates (n = 3 for +, UAS CG7813 17a, and UAS CG7813 J1; n = 4 for UAS CG7813 + ATPsynD RNAi; n = 2 for ATPsynD RNAi and ATPsynG RNAi). Error bars represent 95% confidence intervals. c-d Confocal images of Drosophila flight muscle from Act88F-GAL4; UAS-mitoGFP (c) flies and Act88F-GAL4; UAS-mitoGFP/UAS-CG7813 flies (d). Scale bar 10 μm
To ask whether the Knon-mediated shift toward more ATP synthase monomers was associated with altered flight muscle mitochondrial morphology, we examined by confocal microscopy tissue from flies expressing Knon in flight muscle along with a separate mitochondrially-targed GFP (mitoGFP) [39] (Fig. 4c-d). The mitoGFP localization was roughly similar between flies with and without exogenously expressed Knon, suggesting no gross alteration in mitochondrial number or size. Consistent with the lack of obvious effect on mitochondrial structure, flies expressing Knon in flight muscle were viable and able to fly, though there may be subtle effects at the levels of tissue architecture and flight ability that our experiments did not detect.
Other testis-enriched paralogs of ATP synthase subunits diverged at different times within insect lineages and have roles in spermatid mitochondrial dynamics
In addition to subunit d, the D. melanogaster genome includes pairs of paralogs encoding ATP synthase subunits F6, b, and g (which are in the peripheral stalk), as well as ε and β (in the F1 portion of the complex) [27]. Expression data from modENCODE [30] and FlyAtlas [28] consistently indicate strong testis enrichment for one gene within each paralogous pair. Similar to subunit d, for subunit F6 the testis-enriched predicted gene product (147 amino acids) is significantly larger than that of the broadly-expressed paralog (106 amino acids). Conversely, for subunit b the testis-enriched predicted gene product (152 amino acids) is significantly smaller than that of the broadly expressed paralog (243 amino acids).
We searched other sequenced genomes and found that the genes encoding ATP synthase subunits diverged into paralogous pairs at different times in insect lineages (Fig. 5). Maximum likelihood phylogeny analysis (not shown) confirmed that the proteins encoded by a given pair of paralogs across species clustered in two groups around each of the differentially expressed D. melanogaster paralogs. In each case, the grouping that included the D. melanogaster testis-enriched paralog showed a much higher per-site amino acid substitution rate (Fig. 5b), suggesting that they are evolving faster, consistent with previous observations [8]. For each subunit, the two paralogs were not in close proximity in the genome, and in no case was there synteny in their respective regions. For the paralogs encoding the ATP synthase F6 subunit, the testis-enriched version lacks introns, consistent with retroduplication as a possible duplication mechanism [40]. For subunits b and g, the testis-enriched paralog contains introns but fewer than in the broadly-expressed version, consistent with a possible retroduplication mechanism involving alternatively spliced transcripts as the gene duplication source [41].
Fig. 5.
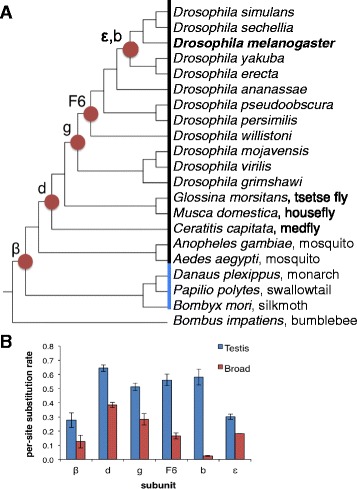
Phylogeny and evolution of Knon and other ATP synthase subunit paralogs in insects. a The six ATP synthase subunits encoded by two paralogs in D. melanogaster have widely varying phylogenetic origins, determined by comparisons among insect genome sequences (black, Diptera; blue, Lepidoptera) deposited in NCBI. Circles indicate the most recent common ancestor that contained both paralogs. b Testis-enriched subunits have acquired more amino acid substitutions relative to a recent outgroup than broadly expressed subunits. The testis-specific ortholog of each subunit was inferred based on sequence similarity to D. melanogaster orthologs, which were in all cases unambiguous. The average per-site amino acid substitution rate for each subunit type relative to the most recent outgroup of the gene duplication was calculated from a Clustal Omega alignment [76]. Error bars indicate standard deviation among the species analyzed
Inducing RNAi knockdown of testis-specific ATP synthase subunits F6 and g in the Drosophila melanogaster testis using the bam-GAL4 driver led to mitochondrial elongation defects similar to those seen in homozygous knon males (Fig. 6), while knockdown of the broadly-expressed paralogs showed more subtle effects. Conversely, knockdown of the more recently diverged testis-specific b subunit did not affect spermatogenesis, while the broadly-expressed subunit b appears to retain an essential role in mitochondrial shaping during spermatogenesis. In contrast to these results, testis knockdown of the sole ortholog of the ATP synthase α subunit (bellwether) [42], which is in the F1 portion and is crucial for ATP synthesis, resulted in degenerating spermatid cysts filled with large, prominent vacuolar inclusions. In general, these results are consistent with the premise that the ATP synthase complex controls high-level mitochondrial architecture in developing spermatids through a mechanism separable from ATP synthesis.
Fig. 6.
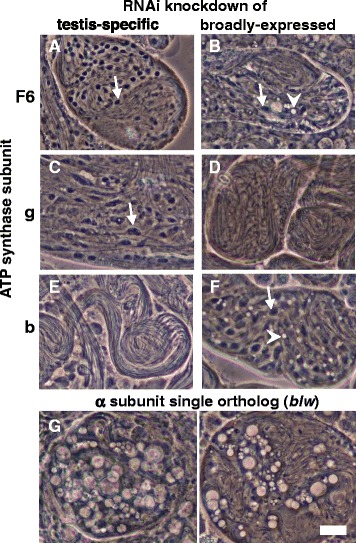
RNAi knockdown of other ATP synthase subunits in the testis. a-f bam-GAL4-driven UAS-RNAi testis knockdown of ATP synthase subunit paralogs that show either testis-enriched expression (left column) or broad expression across multiple tissues (right column). Two of the three testis-enriched paralogs of peripheral stalk subunits—F6 (a) and g (c), but not b (e)—show a similar knockdown phenotype to knon, with failure of mitochondrial elongation (arrows) in spermatid cysts that have nearly fully elongated, along with individualization failure, a common secondary effect of spermatid morphology defects [6]. Knockdown of broadly-expressed paralogs of subunits F6 (b) and b (f) results in aberrant mitochondrial elongation along with vacuolar inclusions (arrowheads), whereas knockdown of broadly-expressed subunit g (d) results in elongating spermatid cysts of wild-type appearance. g Testis knockdown of the sole ATP synthase α subunit ortholog (bellwether), part of the F1 portion of ATP synthase, resulted in large vacuolar inclusions and degradation of elongating spermatid cysts. Two representative cysts are shown. Scale bar 20 μm for a-g. For each genotype, >80 spermatid cysts were visualized from >6 dissected males
Discussion
The Knon ATP synthase subunit d paralog is required for normal Nebenkern morphology
We found a role for Knon, an uncharacteristically large paralog of ATP synthase subunit d, in internal shaping of mitochondrial membranes in the Nebenkern during Drosophila spermatogenesis. Male flies lacking functional Knon were sterile and showed unusual membrane curvature and faulty internal structure within the Nebenkern (Fig. 1), with subsequent entanglement of the two unfurling mitochondrial derivatives and aberrant mitochondrial elongation as the flagellar axoneme grows. Simultaneously preventing mitochondrial fusion in a knon; fzo double mutant suppresses the knon single mutant elongation defect. This result indicates that the knon phenotype is due to the formation of an aberrant Nebernkern structure that cannot elongate, rather than defects at later stages. Knon is expressed only in the testis [28], and the protein is detected on mitochondria from meiosis through spermatid elongation (Fig. 3), consistent with the timing of dramatic mitochondrial membrane reorganization.
The effect of knon on mitochondrial shaping is likely at the level of mitochondrial dynamics and not resulting from respiratory deficiency, since 1) the mitochondrial membrane potential remains high in knon mutant spermatids; 2) other energy-requiring events of spermatid development, such as axoneme elongation and nuclear shaping, proceed; 3) disrupting ATP synthesis by knocking down the sole ATP synthase α subunit ortholog in the testis leads to tissue degeneration; and 4) in other systems, the effects of ATP synthase on organelle morphology and respiration are separable [15, 16].
In many cell types, dimerization of ATP synthase complexes with their axes at an angle of 86° is important for determining sharp positive curvature of the inner mitochondrial membrane at cristae tips. In the Nebenkern, however, the majority of the inner mitochondrial membrane is closely apposed to the outer mitochondrial membrane and shows very shallow curvature. The unusual membrane configuration in the Nebenkern represents accumulation of inner and outer membrane material that is subsequently needed during mitochondrial elongation [20]. We hypothesized that, normally, Knon expression during spermiogenesis may inhibit or alter ATP synthase dimerization to facilitate the more shallow curvature of the bulk of the inner mitochondrial membrane as the Nebenkern forms. In knon mutant spermatids, because the broadly-expressed paralog of subunit d (CG6030) is also expressed in the testis [28, 30], Nebenkern membranes may show aberrant sharp curvature from ATP synthase dimerization (Fig. 7).
Fig. 7.
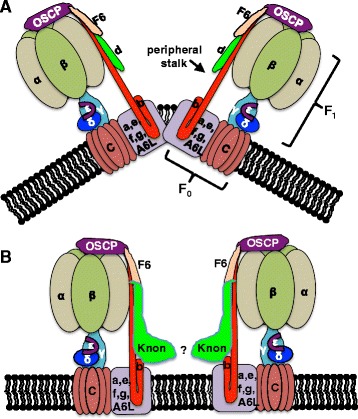
Model for alteration of ATP synthase complex dimerization by Knon. a Schematic representation of ATP synthase complex dimerization as mediated by various subunits in the peripheral stalk and F0 portion. b Steric interactions of the large Knon C termini might inhibit dimerization or enable dimerization at a shallower angle
Effects of Knon on ATP synthase dimerization
In yeast and mammalian cells, ATP synthase subunits e, g, and b are known to mediate dimerization of ATP synthase complexes and contribute to the sharp curvature of inner mitochondrial membranes at cristae tips [43, 44]. S. cerevisiae cells mutant for subunit e or subunit g, normally found in the membrane-embedded F0 portion of the complex, lack higher order ATP synthase dimers and have onion-like mitochondrial structures, reminiscent of the fly Nebenkern, that lack the tight inner membrane curvature normally found in cristae [45–47]. Subunit b in the peripheral stalk is also important for dimerization of the ATP synthase complex, and it can homodimerize on its own [48, 49]. In yeast with a truncated subunit b missing the transmembrane portion, no ATP synthase dimers form, and inner mitochondrial membranes lack cristae [44].
Peripheral stalk components farther away from the membrane also contribute to ATP synthase dimerization. Subunit h (F6 in metazoans), in the peripheral stalk, can homodimerize and stabilize dimers in yeast even if subunits e and g are missing [50, 51]. Cryo-electron microscopy of ATP synthase dimers from the alga Polytomella indicates that peripheral stalk components from adjacent ATP synthase monomers intertwine at a position 80 angstroms away from the inner mitochondrial membrane [52], consistent with the positions of subunits d and h/F6. The broadly-expressed D. melanogaster subunit d (CG6030) has 178 amino acids and is expressed in all tissues, although at lower levels in testes [28]. Orthologs in humans, mice, C. elegans, and budding yeast are similar in size, with 161, 161, 191, and 174 amino acids, respectively. In contrast, Knon has 734 amino acids as a result of a large C-terminal extension. The crystal structure of bovine subunit d suggests that its carboxy terminus extends from the middle of the peripheral stalk toward the membrane and embedded F0 portion [23], consistent with the idea that Knon’s extra large domain in that position may alter the interactions between ATP synthase complex monomers.
Exogenous expression of Knon in flight muscle indeed reduces dimerization of ATP synthase (Fig. 4). Expression of Knon in the absence of testis-specific versions of subunits F6, g, and b may not, however, allow for assembly dynamics of a full testis-specific ATP synthase complex. We therefore cannot rule out the possibility that in wild-type spermatids, Knon along with the other testis-specific paralogs may help mediate ATP synthase dimerization at a shallower angle. Dimerization of ATP synthase complexes at a shallower angle has been seen in Tetrahymena [53]. ATP synthase dimerization in the testis is difficult to assess directly because of tissue heterogeneity and technical constraints.
Truncated KnonMi10492 lacking the carboxy-terminal 180 amino acids is partially functional, suggesting that 554 amino acids (plus any encoded on the inserted element before the first stop codon is encountered), are sufficient for Knon to perform its function when threshold levels of protein are present. These data are consistent with the carboxy-terminal extension either providing steric hindrance that blocks dimerization or promoting unique interactions that underlie dimerization at a different angle.
Roles for other testis-enriched ATP synthase paralogs in spermatid mitochondrial shaping, and implications of different divergence times
Three other subunits of ATP synthase in the peripheral stalk and/or at the putative dimerization interface (F6, g, and b), and two subunits in the F1 portion (β and ε), appear to be encoded by broadly-expressed and testis-specific paralogs in the Drosophila melanogaster genome. We showed via RNAi knockdown experiments that the testis-specific paralogs of subunits F6 and g are required for proper Nebenkern dynamics during spermatogenesis, and that these phenotypes differed from the outcome of disrupting ATP synthesis via knockdown of the sole α subunit ortholog. The testis-enriched F6 subunit, like the Knon subunit d, is larger than the broadly-expressed version, while the two g subunit paralogs are similar in size. The testis-enriched g subunit therefore might not provide a steric hindrance to dimerization of the complex but might instead promote assembly of the testis-enriched large d and F6 subunits into the complex, or promote interactions between adjacent ATP synthase complexes at a shallower angle at the base of the peripheral stalk.
For subunit b, the RNAi transgenic line targeting the testis-enriched paralog may not be sufficiently inducible to show an effect, or alternatively the testis-enriched subunit b might truly be not required for spermatid morphogenesis. Consistent with our data, another group recently showed that knockdown of the broadly-expressed ATPsynB results in male infertility and a lack of mature sperm, though they did not examine the mitochondrial phenotype [54]. Dispensability of the testis-specific subunit b would be consistent with its relatively recent gene duplication (Fig. 5), such that there has been less time for specialization of the two paralogs compared to the other paralogous pairs.
The mechanism of differential gene regulation between the paralogs of peripheral stalk ATP synthase subunits is unknown. Notably, the paralogous ATP synthase β subunits were duplicated earlier than the others, and a recent study showed that the broadly-expressed Drosophila ATPsynβ is normally repressed in germ cells by the hairpin RNA pathway [55]. Derepression of ATPsynβ via knockout of the appropriate hpRNA causes defects in germ cell differentiation and male sterility [55]. These findings suggest that the ATPsynβ paralogs have functionally diverged and have evolved RNA-based regulatory mechanisms to favor the expression of one paralog over the other in the testis. The mechanisms of regulation of the other paralogous pairs are yet to be determined.
ATP synthase, mitochondrial shaping, and tissue differentiation
In mammalian systems the ultrastructural organization of the inner mitochondrial membrane can vary among tissue types and in response to metabolic state or developmental cues (reviewed in [14]). Expression of subunit e, important for dimerization, can be triggered by altered nutrient and oxygen levels [56, 57]. In differentiating cardiomyocytes, mitochondria undergo structural changes concomitant with ATP synthase dimer stabilization by an accessory factor [58, 59]. In the syncytiotrophoblast outer layer of the placenta, reduced ATP synthase dimerization accompanies a lack of cristae, despite normal ATP production [60]. Specific mutations in the mitochondrially-encoded ATP6 subunit found in Leigh syndrome patients are associated with higher levels of ATP synthase dimerization and decreased turnover of the complex [61].
The Drosophila ovary requires broadly-expressed ATP synthase components for germ cell differentiation [62], possibly associated with mitochondrial morphology and not respiration, because knockdowns of other respiratory complexes did not show the same effect. Germline-specific ATP synthase subunits in C. elegans appear required for tissue differentiation [63], though no mitochondrial structural aspects were examined. In trypanosomes, different ATP synthase subunit c paralogs are encoded in the genome; it is not yet known whether these gene products influence mitochondrial function and morphology in different life cycle stages [64], though subunit c is not positioned to influence dimerization of the complex directly. Our findings in the Drosophila testis therefore elucidate the first known instance in which tissue-specific ATP synthase subunits underlie tissue-specific regulation of mitochondrial shaping.
Conclusions
We describe the first evidence connecting tissue-specific mitochondrial shaping with the expression and function of tissue-specific ATP synthase subunits. A testis-specific variant of ATP synthase subunit d in Drosophila is required for Nebenkern morphology and appears to alter dimerization of ATP synthase complexes. Testis-specific paralogs of other ATP synthase subunits also appear to have a role. Our data, showing the role of a cell biological process in the context of tissue morphogenesis, contribute to the growing knowledge of ATP synthase’s role in mediating membrane conformation, beyond its canonical role in ATP production. Future work with tissue-specific ATP synthase subunit paralogs in Drosophila may explore their sub-mitochondrial localization to test localization in areas of shallower inner mitochondrial membrane curvature; further work should also explore whether the testis-specific paralogs together form a unique complex, and whether individual subunits can be substituted between the different forms of the complex.
Methods
Fly stocks, husbandry, and fertility tests
Flies were raised on standard cornmeal molasses medium or instant potato flake medium (Ward’s Scientific) at 25 °C for all crosses except for those combining GAL4 and UAS transgenes, which were maintained at 29 °C. Multiply marked stocks, transposable element insertions, the Δ2-3 transposase-encoding stock, the Act88F-GAL4 driver stock, and all deficiencies except Df(2R)7E were obtained from the Bloomington Drosophila Stock Center. Oregon R was used as the wild-type strain. Flies with transgenic UAS-RNAi constructs were obtained from the Vienna Drosophila Resource Center. The bam-GAL4 driver stock was a gift from Yukiko Yamashita. Most fertility tests were performed by mating individual males with three Oregon R virgin females and assessing the presence of larvae after five days. For quantifying fertility of knon Mi10492 allelic combinations, 56 age-matched males of each genotype were placed individually in vials with three age-matched Oregon R virgin females; pupae and eclosed offspring from vials in which the males survived were counted after fifteen days. Data were analyzed by T test.
RNAi knockdown
We obtained all RNAi transgenic lines from the Vienna Drosophila Resource Center [32], listed here according to their targets:
P[GD12291]v22566 (knon; CG7813; testis-specific subunit d)
P[KK1096241]v104818 (CG12027; testis-enriched subunit F6)
P[KK108581]v107826 (CG4412; broadly-expressed subunit F6)
P[GD12493]v35387 (CG7211; testis-enriched subunit g)
P[KK108323]v107311 (CG6105; broadly-expressed subunit g)
P[GD1688]v6521 (CG17300; testis-enriched subunit b)
P[GD6129]v14211 (CG8189; broadly-expressed subunit b)
P[GD11030]v34663 (CG3612 (bellwether); sole ortholog of subunit α)
For knockdown in the testis, each RNAi line was crossed to a bam-GAL4 driver line [31]. Testes of F1 males were dissected and imaged as described below. For RNAi knockdown in flight muscle, each relevant UAS-RNAi line was crossed to the Act88F-GAL4 third chromosome driver line (Bloomington Drosophila Stock Center).
Microscopy
For light microscopy, testes were dissected in TB1 buffer (7 mM K2HPO4, 7 mM KH2PO4 (pH 6.7), 80 mM KCl, 16 mM NaCl, 5 mM MgCl2, 1% PEG-6000) and opened with forceps to allow cells to form a monolayer under pressure of a cover slip. Co-labeling of DNA and polarized mitochondria was performed by incubating intact testes in TB1 supplemented with 5% DMSO, 2.5 μg/mL Hoechst, and 10 μg/mL rhodamine 123 [65] for ~10-15 min in the dark. The testes were washed three times with TB1 to remove residual stain and then torn open on a polylysine-coated slide prior to imaging. Phase-contrast and epifluorescence microscopy were performed on a Nikon Eclipse E600W, Olympus B201, or Olympus BX60 microscope, and images were captured with a Nikon Coolpix 4500 or Olympus DP80 camera.
For confocal microscopy, thoraces of adult males and females were dissected to obtain flight muscle tissue. Thoraces were fixed in 4% paraformaldehyde with PBS, washed 3x with PBT (0.2% TritonX), and stained overnight with tetramethylrhodamine-conjugated phalloidin in PBT at a concentration of 8 μM. Whole thoraces were mounted on a slide with mounting media and sealed with a coverslip and clear nail polish. The flight muscle extruded from the exoskeleton after pressing and sealing the coverslip. Samples were observed with a Nikon 90i confocal microscope and a Plan Apo TIRF 100X/1.45 oil objective. Images were captured and overlaid using the confocal software EZ-C1.
For transmission electron microscopy, testes were dissected in TB1 and immediately placed in fixative (2% glutaraldehyde, 1% paraformaldehyde, 0.1 M sodium phosphate or sodium cacodylate buffer (pH 7)). After overnight fixation, samples were washed in 0.1% phosphate or cacodylate buffer for 15 min and stained with 1% osmium tetroxide in the same buffer for two hours. Testes were then washed three times in water, stained for 1 h in 1% uranyl acetate, washed three times in water and dehydrated through an ethanol series (30%, 50%, 70%, 95%, 100%). After five minutes in 1:1 ethanol:propylene oxide and five minutes in propylene oxide, samples were embedded in Spurr’s resin and polymerized overnight at 60 °C. Thin sections (80-90 nm) were cut with a Reichert-Jung microtome, placed on Formvar-coated slot grids and examined on a Phillips 410 transmission electron microscope.
Recombination and deficiency mapping
Virgin females of genotype knon/ al dp b r cn vg c a px bw mi sp were crossed to al dp b pr Bl c px sp/CyO males. F1 males that carried Bl along with other markers indicating single recombination events on the second chromosome were selected for each interval and crossed individually to w; knon/CyO females to score both knon and the w +-marked P element, and to make stocks of the recombinant chromosome with CyO. For finer mapping, knon/c wt px females were crossed to al dp b pr Bl c px sp/CyO males, and F1 male recombinant progeny in the c-px interval were individually crossed to knon/CyO to score knon and make stocks of recombinant chromosomes. The welt (wt) marker was scored in homozygotes in each of the recombinant stocks. For deficiency mapping, knon/CyO flies were crossed to selected 2nd chromosome deficiency lines balanced over CyO, and the Cy+ male offspring were tested for fertility and dissected as described above to assess testis subcellular phenotype.
Generation of transgenic lines
A knon transgene with endogenous regulatory sequences was generated by amplifying a ~5 kb genomic region containing CG7813 along with ~1.8 kb upstream and ~0.5 kb downstream, with primers 5’ ATATATGGTACCGCCATCAGCCAGCGAGCTT 3’ (forward) and 5’ GCATACCGCGGGGCAACACTTTTCGAACGG 3’ (reverse), and cloning the fragment into the pCaSpeR4 vector [66] using restriction sites KpnI and SacII. Primers were purchased from Integrated DNA Technologies (Coralville, IA). A separate knon-GFP transgene was created to include the ~1.8 kb upstream region and knon coding region through the penultimate codon, with GFP (GFPmut3* variant with S2R, S65G, S72A [67]) ligated in-frame 3’ to the coding region at the SacII restriction site. To create UAS-knon and UAS-knon-GFP constructs, the knon and knon-GFP constructs described above were used as PCR template in two-step splicing by overlap extension. The single knon intron was removed by amplifying the two exons of the gene in separate PCR reactions, using Phusion High-Fidelity PCR Master Mix with HF Buffer (New England Biolabs Mo531S). The two products were then pooled in a third reaction, where they annealed and amplified to form a product with sequence identical to the cDNA. The inserts were cloned into pUAST ([38]; obtained from the Drosophila Genomics Resource Center, Bloomington, IN) using KpnI and XbaI (New England Biolabs). DNA sequencing to check constructs was performed by Retrogen (San Diego, CA), and DNA sequence analysis was performed in ApE [68]. Constructs were injected into w 1118 embryos by Rainbow Transgenics (Camarillo, CA). w + F1 offspring were outcrossed to Kr/CyO; D/TM6C to make stocks and map insertions. knon and knon-GFP transgenes inserted on the third chromosome were crossed into the knon mutant background.
BN-PAGE and immunoblotting of flight muscle mitochondria
Mitochondria were extracted from fly thoraces using a protocol modified from [69]. Fifteen thoraces from flies of each genotype were crushed in 50 μL mitochondrial isolation medium (MIM: 250 mM sucrose, 10 mM Tris-HCl, pH 7.4, 0.15 mM MgCl2) using a plastic pestle, then spun twice at 500 x g for 5 min at 4 °C to remove debris. The supernatant was spun at 5,000 x g for 5 min at 4 °C to pellet mitochondria. The pellet was subjected to the Life Technologies NativePAGE Novex® Bis-Tris Gel System protein isolation protocol for BN-PAGE. Protein pellets were resuspended in 25 μL of 1x Native PAGE sample buffer (Invitrogen) containing 1% digitonin. Samples were incubated on ice for 15 min and centrifuged at 20,000 x g for 30 min at 4 °C. The supernatant was frozen at -80 °C. Using reagents and protocol from Life Technologies, NativePAGE Novex® Bis-Tris Gel System, G-250 dye was added to samples to a final concentration of 0.25%, and samples were run on a 3-12% polyacrylamide gel for 90 min at 150 V constant in the X-Cell Sure Lock Mini Cell gel apparatus by Life Technologies. Immunoblotting was performed with the X Cell II Blot Module (Invitrogen). The gel was soaked for 15 min in a 0.1% SDS solution with water and samples transferred to PVDF membrane in the X Cell transfer apparatus on ice for 1 h. Following transfer to the membrane, the membrane was fixed in 8% acetic acid and rinsed with methanol and dH2O. The primary antibody was mouse monoclonal anti-ATP5A antibody [15H4C4] by Abcam at a dilution of 1:3000 or 1:5000. The blocking buffer was StartingBlock (PBS) Blocking Buffer from Thermo Scientific with 0.1% Tween. The secondary antibody was goat anti-mouse IgG-HRP by Santa Cruz Biotechnology [sc-2005] at a dilution of 1:2500. Primary incubation occurred overnight, shaking at 4 °C and secondary incubation occurred at RT, shaking for 1 h. Antibodies were diluted with blocking buffer without Tween. Membranes were imaged and band intensity quantified using a Bio-Rad gel imager for chemiluminescence and ImageJ software. To determine the internal ratio of the dimer to monomer bands within each given lane, background was subtracted and band intensity measured on non-saturated blot images.
Phylogenetic analysis
ATP synthase subunit paralogs were identified through FlyBase annotations and orthology tools [27, 70] supplemented with NCBI BLAST [71] and VectorBase [72] to search for ATP synthase subunits encoded by multiple paralogs in genomes outside the Drosophila genus. We used maximum likelihood phylogeny [73] to test if the proteins encoded by all paralogs cluster into two distinct groups. Clustering, tree drawing, and evolution rate analyses were performed in the MEGA5 suite [74, 75].
Acknowledgements
We thank Patricia Wilson for the knon ms(2)1400 allele, Bee Choo Liang and Anthony Mahowald for help with initial mapping and Fran Thomas for assistance with electron microscopy. The Davidson College Fall 2009 Genetics class assisted with deficiency mapping of knon ms(2)1400. Sarah Pyfrom and Ben Jepson provided technical assistance. We thank Barbara Lom for valuable guidance with the confocal microscope, and Jon Lim and Riley Mangan for assistance with immunoblots. Many thanks to three anonymous reviewers and Devon Harris for constructive suggestions on the manuscript. FlyBase, the Bloomington Drosophila Stock Center, and the Vienna Drosophila Resource Center were invaluable resources for data and fly stocks.
Funding
Work in the Hales lab was supported by National Science Foundation RUI 1158024 as well as by the Davidson College Department of Biology. YH, COF, and OM were supported by Davidson Research Initiative grants. In the mid-1990s, when this project was initiated, MTF was funded by National Institutes of Health (NIH) grant 1RO1-HD29194, and KGH was funded by a Howard Hughes Medical Institute predoctoral fellowship.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
MTF and KGH conceptualized, respectively, initial and later parts of this project. EMS, ECB, YH, LEI, OB, MB, DA, KES, OM, COF, NS, MGG, ETG, LAR, CGW, and KGH designed experiments and acquired, analyzed, and interpreted data. KGH drafted the manuscript with substantial contributions from EMS and ECB, and MTF also provided editorial contributions. This work was begun in the 1990s when KGH was a predoctoral student in the laboratory of MTF, and the bulk of the work was later completed after a long hiatus, in the laboratory of KGH. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable
Ethics approval and consent to participate
Not applicable
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eric M. Sawyer, Email: sawyer.ericm@gmail.com
Elizabeth C. Brunner, Email: lizbrunner18@gmail.com
Yihharn Hwang, Email: yihharn@gmail.com.
Lauren E. Ivey, Email: laivey89@gmail.com
Olivia Brown, Email: olivia.brown2019@gmail.com.
Megan Bannon, Email: megbann@gmail.com.
Dennis Akrobetu, Email: deakrobetu@davidson.edu.
Kelsey E. Sheaffer, Email: kesheaffer@gmail.com
Oshauna Morgan, Email: oshaunamorgan@gmail.com.
Conroy O. Field, Email: cofield@udel.edu
Nishita Suresh, Email: nishita.suresh@gmail.com.
M. Grace Gordon, Email: m.gracie.gordon@gmail.com.
E. Taylor Gunnell, Email: tagunnell@hotmail.com.
Lindsay A. Regruto, Email: lindsay056@gmail.com
Cricket G. Wood, Email: c_wood@lifesci.ucsb.edu
Margaret T. Fuller, Email: mtfuller@stanford.edu
Karen G. Hales, Email: kahales@davidson.edu
References
- 1.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/S0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 4.Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/S0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- 5.Tates AD. Cytodifferentiation during spermatogenesis in Drosophila melanogaster: an electron microscope study. Leiden: Rijksuniversiteit; 1971. [Google Scholar]
- 6.Fuller MT. Spermatogenesis. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor: Cold Spring Harbor Press; 1993. pp. 71–147. [Google Scholar]
- 7.Fabian L, Brill JA. Drosophila spermiogenesis: big things come from little packages. Spermatogenesis. 2012;2:197–212. doi: 10.4161/spmg.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assis R, Bachtrog D. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci U S A. 2013;110:17409–17414. doi: 10.1073/pnas.1313759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devenish RJ, Prescott M, Rodgers AJ. The structure and function of mitochondrial F1F0-ATP synthases. Int Rev Cell Mol Biol. 2008;267:1–58. doi: 10.1016/S1937-6448(08)00601-1. [DOI] [PubMed] [Google Scholar]
- 10.Kucharczyk R, Zick M, Bietenhader M, Rak M, Couplan E, Blondel M, Caubet SD, di Rago JP. Mitochondrial ATP synthase disorders: molecular mechanisms and the quest for curative therapeutic approaches. Biochim Biophys Acta. 2009;1793:186–199. doi: 10.1016/j.bbamcr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Habersetzer J, Ziani W, Larrieu I, Stines-Chaumeil C, Giraud MF, Brethes D, Dautant A, Paumard P. ATP synthase oligomerization: from the enzyme models to the mitochondrial morphology. Int J Biochem Cell Biol. 2013;45:99–105. doi: 10.1016/j.biocel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Seelert H, Dencher NA. ATP synthase superassemblies in animals and plants: two or more are better. Biochim Biophys Acta. 1807;2011:1185–1197. doi: 10.1016/j.bbabio.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Wittig I, Schagger H. Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim Biophys Acta. 2009;1787:672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Zick M, Rabl R, Reichert AS. Cristae formation—linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19. [DOI] [PubMed]
- 15.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schagger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forster K, Turina P, Drepper F, Haehnel W, Fischer S, Graber P, Petersen J. Proton transport coupled ATP synthesis by the purified yeast H+ -ATP synthase in proteoliposomes. Biochim Biophys Acta. 2010;1797:1828–1837. doi: 10.1016/j.bbabio.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Baker LA, Watt IN, Runswick MJ, Walker JE, Rubinstein JL. Arrangement of subunits in intact mammalian mitochondrial ATP synthase determined by cryo-EM. Proc Natl Acad Sci U S A. 2012;109:11675–11680. doi: 10.1073/pnas.1204935109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kuhlbrandt W. Structure of the yeast F1F0-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci U S A. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strauss M, Hofhaus G, Schroder RR, Kuhlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokuyasu KT. Dynamics of spermiogenesis in Drosophila melanogaster. VI. Significance of "onion" nebenkern formation. J Ultrastruct Res. 1975;53:93–112. doi: 10.1016/S0022-5320(75)80089-X. [DOI] [PubMed] [Google Scholar]
- 21.Walker JE, Runswick MJ, Poulter L. ATP synthase from bovine mitochondria. The characterization and sequence analysis of two membrane-associated sub-units and of the corresponding cDNAs. J Mol Biol. 1987;197:89–100. doi: 10.1016/0022-2836(87)90611-5. [DOI] [PubMed] [Google Scholar]
- 22.Norais N, Prome D, Velours J. ATP synthase of yeast mitochondria. Characterization of subunit d and sequence analysis of the structural gene ATP7. J Biol Chem. 1991;266:16541–16549. [PubMed] [Google Scholar]
- 23.Bueler SA, Rubinstein JL. Location of subunit d in the peripheral stalk of the ATP synthase from Saccharomyces cerevisiae. Biochemistry. 2008;47:11804–11810. doi: 10.1021/bi801665x. [DOI] [PubMed] [Google Scholar]
- 24.Dickson VK, Silvester JA, Fearnley IM, Leslie AG, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson PG, Fuller MT, Borisy GG. Monastral bipolar spindles: implications for dynamic centrosome organization. J Cell Sci. 1997;110(Pt 4):451–464. doi: 10.1242/jcs.110.4.451. [DOI] [PubMed] [Google Scholar]
- 26.Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13:R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM, FlyBase C. FlyBase: introduction of the Drosophila melanogaster release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20. [DOI] [PubMed]
- 29.Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75:1389–1399. doi: 10.1016/0092-8674(93)90625-Z. [DOI] [PubMed] [Google Scholar]
- 30.modEncode Consortium. Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D, McKearin DM. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 2003;130:1159–1170. doi: 10.1242/dev.00325. [DOI] [PubMed] [Google Scholar]
- 32.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 33.Nagarkar-Jaiswal S, Lee PT, Campbell ME, Chen K, Anguiano-Zarate S, Gutierrez MC, Busby T, Lin WW, He Y, Schulze KL et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife. 2015;4:e05338. [DOI] [PMC free article] [PubMed]
- 34.Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nature Methods. 2011;8:737–43. [DOI] [PMC free article] [PubMed]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 37.White-Cooper H, Schafer MA, Alphey LS, Fuller MT. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development. 1998;125:125–134. doi: 10.1242/dev.125.1.125. [DOI] [PubMed] [Google Scholar]
- 38.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 39.Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/S0960-9822(95)00128-X. [DOI] [PubMed] [Google Scholar]
- 40.Diaz-Castillo C, Ranz JM. Nuclear chromosome dynamics in the Drosophila male germ line contribute to the nonrandom genomic distribution of retrogenes. Mol Biol Evol. 2012;29:2105–2108. doi: 10.1093/molbev/mss096. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C, Gschwend AR, Ouyang Y, Long M. Evolution of gene structural complexity: an alternative-splicing-based model accounts for intron-containing retrogenes. Plant Physiol. 2014;165:412–423. doi: 10.1104/pp.113.231696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs H, Stratmann R, Lehner C. A screen for lethal mutations in the chromosomal region 59AB suggests that bellwether encodes the alpha subunit of the mitochondrial ATP synthase in Drosophila melanogaster. Mol Gen Genet. 1998;259:383–387. doi: 10.1007/s004380050826. [DOI] [PubMed] [Google Scholar]
- 43.Arselin G, Vaillier J, Salin B, Schaeffer J, Giraud MF, Dautant A, Brethes D, Velours J. The modulation in subunits e and g amounts of yeast ATP synthase modifies mitochondrial cristae morphology. J Biol Chem. 2004;279:40392–40399. doi: 10.1074/jbc.M404316200. [DOI] [PubMed] [Google Scholar]
- 44.Soubannier V, Vaillier J, Paumard P, Coulary B, Schaeffer J, Velours J. In the absence of the first membrane-spanning segment of subunit 4(b), the yeast ATP synthase is functional but does not dimerize or oligomerize. J Biol Chem. 2002;277:10739–10745. doi: 10.1074/jbc.M111882200. [DOI] [PubMed] [Google Scholar]
- 45.Giraud MF, Paumard P, Soubannier V, Vaillier J, Arselin G, Salin B, Schaeffer J, Brethes D, di Rago JP, Velours J. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim Biophys Acta. 2002;1555:174–180. doi: 10.1016/S0005-2728(02)00274-8. [DOI] [PubMed] [Google Scholar]
- 46.Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velours J, Dautant A, Salin B, Sagot I, Brethes D. Mitochondrial F1F0-ATP synthase and organellar internal architecture. Int J Biochem Cell Biol. 2009;41:1783–1789. doi: 10.1016/j.biocel.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 48.Spannagel C, Vaillier J, Arselin G, Graves PV, Grandier-Vazeille X, Velours J. Evidence of a subunit 4 (subunit b) dimer in favor of the proximity of ATP synthase complexes in yeast inner mitochondrial membrane. Biochim Biophys Acta. 1998;1414:260–264. doi: 10.1016/S0005-2736(98)00174-6. [DOI] [PubMed] [Google Scholar]
- 49.Gavin PD, Prescott M, Devenish RJ. F1F0-ATP synthase complex interactions in vivo can occur in the absence of the dimer specific subunit e. J Bioenerg Biomembr. 2005;37:55–66. doi: 10.1007/s10863-005-4128-8. [DOI] [PubMed] [Google Scholar]
- 50.Fronzes R, Weimann T, Vaillier J, Velours J, Brethes D. The peripheral stalk participates in the yeast ATP synthase dimerization independently of e and g subunits. Biochemistry. 2006;45:6715–6723. doi: 10.1021/bi0601407. [DOI] [PubMed] [Google Scholar]
- 51.Fronzes R, Chaignepain S, Bathany K, Giraud MF, Arselin G, Schmitter JM, Dautant A, Velours J, Brethes D. Topological and functional study of subunit h of the F1F0-ATP synthase complex in yeast Saccharomyces cerevisiae. Biochemistry. 2003;42:12038–12049. doi: 10.1021/bi035270j. [DOI] [PubMed] [Google Scholar]
- 52.Allegretti M, Klusch N, Mills DJ, Vonck J, Kuhlbrandt W, Davies KM. Horizontal membrane-intrinsic alpha-helices in the stator a-subunit of an F-type ATP synthase. Nature. 2015;521:237–240. doi: 10.1038/nature14185. [DOI] [PubMed] [Google Scholar]
- 53.Balabaskaran Nina P, Dudkina NV, Kane LA, van Eyk JE, Boekema EJ, Mather MW, Vaidya AB. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol. 2010;8:e1000418. doi: 10.1371/journal.pbio.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen YN, Wu CH, Zheng Y, Li JJ, Wang JL, Wang YF. Knockdown of ATPsyn-b caused larval growth defect and male infertility in Drosophila. Archive Insect Biochem Physiol. 2015;88:144–54. [DOI] [PubMed]
- 55.Wen J, Duan H, Bejarano F, Okamura K, Fabian L, Brill JA, Bortolamiol-Becet D, Martin R, Ruby JG, Lai EC. Adaptive regulation of testis gene expression and control of male fertility by the Drosophila harpin RNA pathway. Molecular Cell. 2015;57:165–78. [DOI] [PMC free article] [PubMed]
- 56.Levy FH, Kelly DP. Regulation of ATP synthase subunit e gene expression by hypoxia: cell differentiation stage-specific control. Am J Physiol. 1997;272:C457–465. [DOI] [PubMed]
- 57.Swartz DA, Park EI, Visek WJ, Kaput J. The e subunit gene of murine F1F0-ATP synthase. Genomic sequence, chromosomal mapping, and diet regulation. J Biol Chem. 1996;271:20942–20948. doi: 10.1074/jbc.271.34.20942. [DOI] [PubMed] [Google Scholar]
- 58.Bisetto E, Comelli M, Salzano AM, Picotti P, Scaloni A, Lippe G, Mavelli I. Proteomic analysis of F1F0-ATP synthase super-assembly in mitochondria of cardiomyoblasts undergoing differentiation to the cardiac lineage. Biochim Biophys Acta. 1827;2013:807–816. doi: 10.1016/j.bbabio.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 59.Campanella M, Casswell E, Chong S, Farah Z, Wieckowski MR, Abramov AY, Tinker A, Duchen MR. Regulation of mitochondrial structure and function by the F1F0-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 60.De los Rios Castillo D, Zarco-Zavala M, Olvera-Sanchez S, Pardo JP, Juarez O, Martinez F, Mendoza-Hernandez G, Garcia-Trejo JJ, Flores-Herrera O. Atypical cristae morphology of human syncytiotrophoblast mitochondria: role for complex V. J Biol Chem. 2011;286:23911–23919. doi: 10.1074/jbc.M111.252056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cortes-Hernandez P, Vazquez-Memije ME, Garcia JJ. ATP6 homoplasmic mutations inhibit and destabilize the human F1F0-ATP synthase without preventing enzyme assembly and oligomerization. J Biol Chem. 2007;282:1051–1058. doi: 10.1074/jbc.M606828200. [DOI] [PubMed] [Google Scholar]
- 62.Teixeira FK, Sanchez CG, Hurd TR, Seifert JR, Czech B, Preall JB, Hannon GJ, Lehmann R. ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation. Nat Cell Biol. 2015;17:689–696. doi: 10.1038/ncb3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawasaki I, Hanazawa M, Gengyo-Ando K, Mitani S, Maruyama I, Iino Y. ASB-1, a germline-specific isoform of mitochondrial ATP synthase b subunit, is required to maintain the rate of germline development in Caenorhabditis elegans. Mech Dev. 2007;124:237–51. [DOI] [PubMed]
- 64.Gulde PE, Christen L, Brown SV, Williams N. Three distinct isoforms of ATP synthase subunit c are expressed in T. brucei and assembled into the mitochondrial ATP synthase complex. PLoS One. 2013;8:e54039. doi: 10.1371/journal.pone.0054039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson LV, Walsh ML, Chen LB. Localization of mitochondria in living cells with rhodamine 123. Proc Natl Acad Sci U S A. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thummel C, Pirrotta V. Technical notes: new pCaSpeR P element vectors. Dros Inf Serv. 1992;71:150. [Google Scholar]
- 67.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 68.Davis MW. ApE: a plasmid editor. http://biologylabs.utah.edu/jorgensen/wayned/ape/. Accessed 27 Aug 2012.
- 69.Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drosophila 12 Genomes Consortium, Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–18. [DOI] [PubMed]
- 71.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, Hughes DS, Koscielny G, Louis C, Maccallum RM, Redmond SN, et al. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 2012;40:D729–734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30:1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- 76.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


