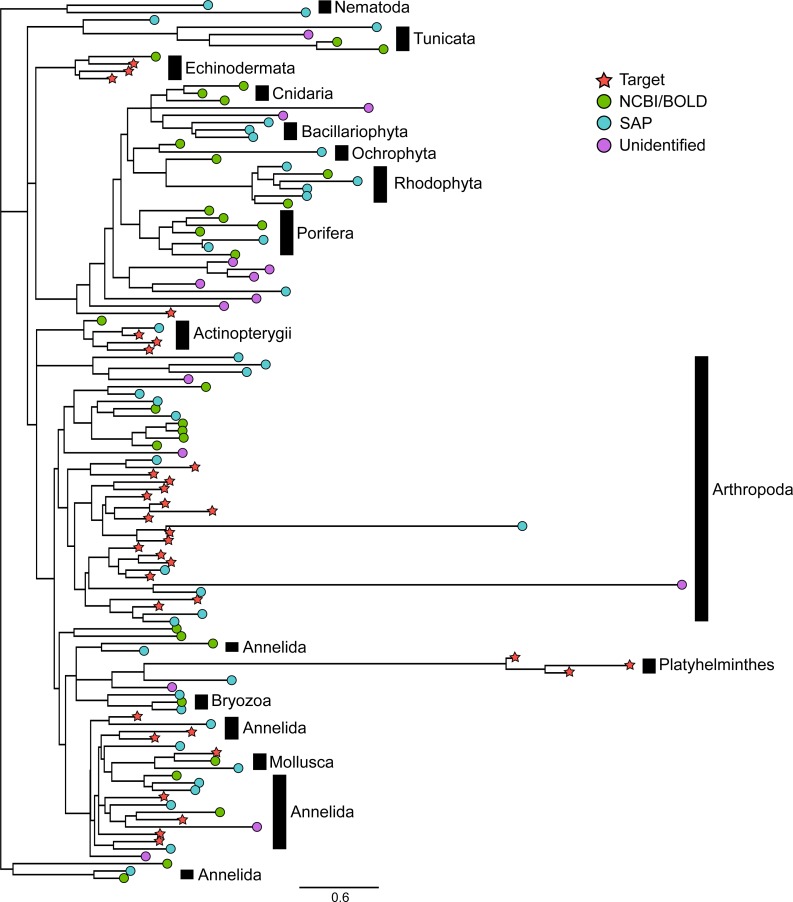
Figure 2. Phylogenetic relationships between representative COI sequences (313 bp) of 120 OTUs detected in the mock sample.
This is the best tree of 1,000 maximum likelihood search replicates computed using the adaptive best tree search analyses implemented in GARLI v2.1 (Zwickl, 2006) through the GARLI web service (Bazinet, Zwickl & Cummings, 2014) . The full tree is provided in Fig. S1. Branch tip symbols indicate the method of identification of each OTU. Target: OTU that matched the COI sequence of a species included in the mock sample (referred to as “target OTUs” throughout the manuscript; note that OTUs that did not match any target OTU are referred to as “non-target OTUs” throughout the manuscript); NCBI/BOLD, OTU that did not match a target OTU but had >97% similarity with a reference COI barcode in NCBI or BOLD; SAP, OTU that did not match a target OTU nor a reference COI barcode in NCBI or BOLD but could be confidently assigned to higher a taxonomic level using a Bayesian phylogenetic approach implemented in the Statistical Assignment Package (SAP); Unidentified, OTU that could not be confidently identified to any taxonomic group using the three approaches detailed above.