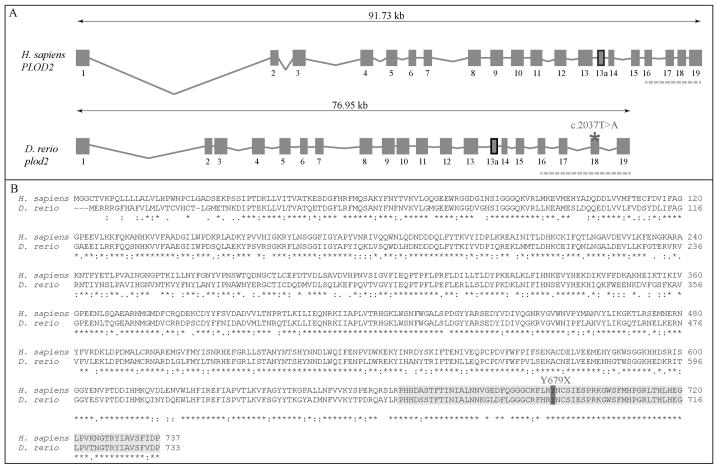
Figure 1. Evolutionary conservation of the zebrafish and human plod2/PLOD2 gene and lh2/LH2 protein.
A. Schematic representation of the genomic structure of the H. Sapiens PLOD2 and D. Rerio plod2 genes. Boxes and lines represent exons and introns of the PLOD2/plod2 genes, respectively. Exon 13a (thick box border) is subject to differential splicing. The catalytic domain is encoded by the last four exons of the PLOD2/plod2 genes (indicated by the dashed line). The asterisk shows the position of the mutation (c.2037T>A) in the plod2sa1768 mutant zebrafish. B. Amino acid sequence alignment (with Clustal Omega) between H. Sapiens LH2 and D. Rerio lh2 (long isoform). The catalytic domain is highlighted in light grey and the mutated residue (p.Y679X) in the plod2sa1768 mutant is indicated in dark grey. Conservation of amino acid sequences are shown below the alignment: “*” means residues identical in all sequences in the alignment; “:” means conserved substitutions; “.” means semi-conserved substitutions; space means no conservation.