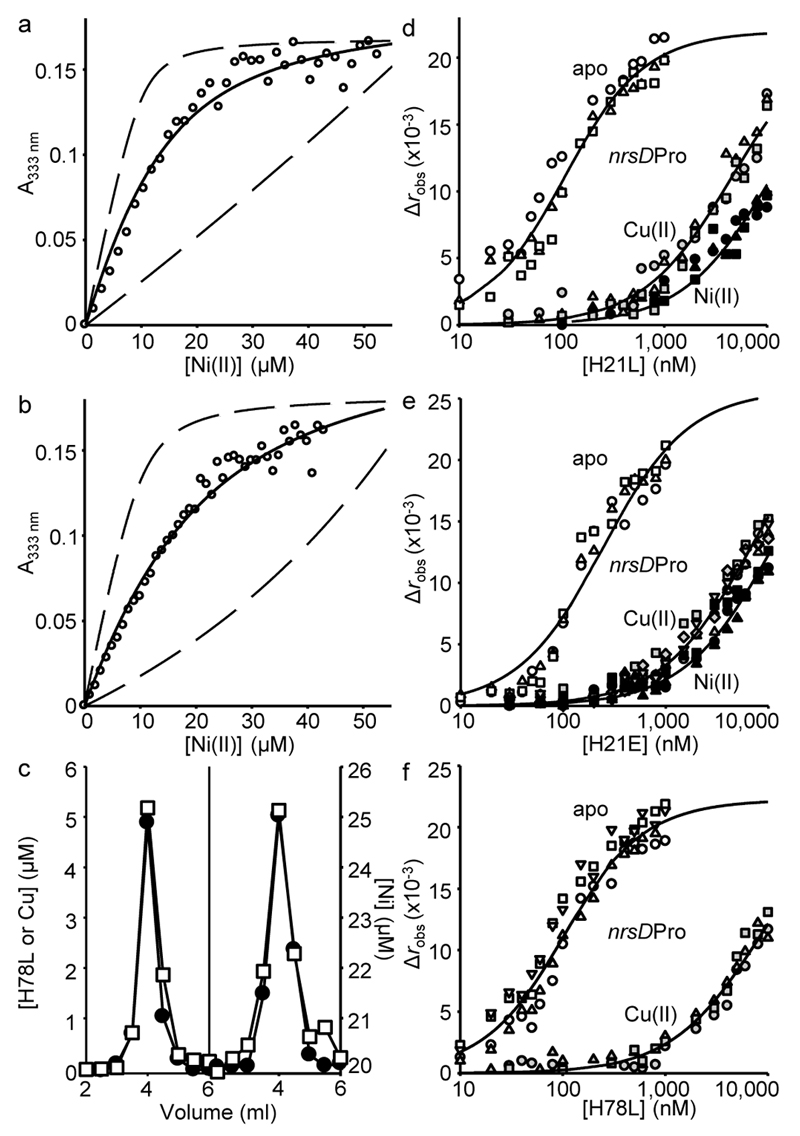
Figure 2. Weak Ni(II)-affinity sensors with altered predicted sensitivities.
a. Representative (n = 3) H21L absorbance upon titration of H21L (10 μM protomer) and EGTA (50 μM) with NiCl2. Solid line is a fit to a model describing competition from H21L for one molar equivalent of Ni(II), revealing KNi(II)= 5.5(±1) × 10–11 M. Dashed lines are simulated curves with KNi(II) 10-fold tighter and 10-fold weaker than the fitted value. b. As in ‘a’ with H21E ([H21E] = 10 μM protomer, [EGTA] = 50 μM), KNi(II) = 8.6(±2) × 10–11 M (n = 3). c. Elution profile of H78L (10 μM protomer) incubated with CuCl2 (15 μM) before (left), or with NiCl2 (20 μM) during (right), fractionation by size exclusion chromatography. Protein (closed symbols), metal (open symbols). Note the 20 μM [Ni] baseline when NiCl2 was included in the buffer. d. Anisotropy change upon titration of nrsDProFA (10 nM) with either H21L in 5 mM EDTA (open symbols), Ni(II)-H21L (closed symbols) or Cu(II)-H21L (gray symbols). Symbol shapes represent individual experiments (n = 3). Data were fit to a model describing a 1:1 InrS tetramer (non-dissociable):DNA stoichiometry and lines are simulated curves using the mean KDNA across the experiments shown (Supplementary Table 2). e. As in ‘d’ with H21E (apo n = 3, Cu(II) n = 3, Ni(II) n = 5). f. As in ‘d’ with H78L (Cu(II) n = 3, apo n = 4).