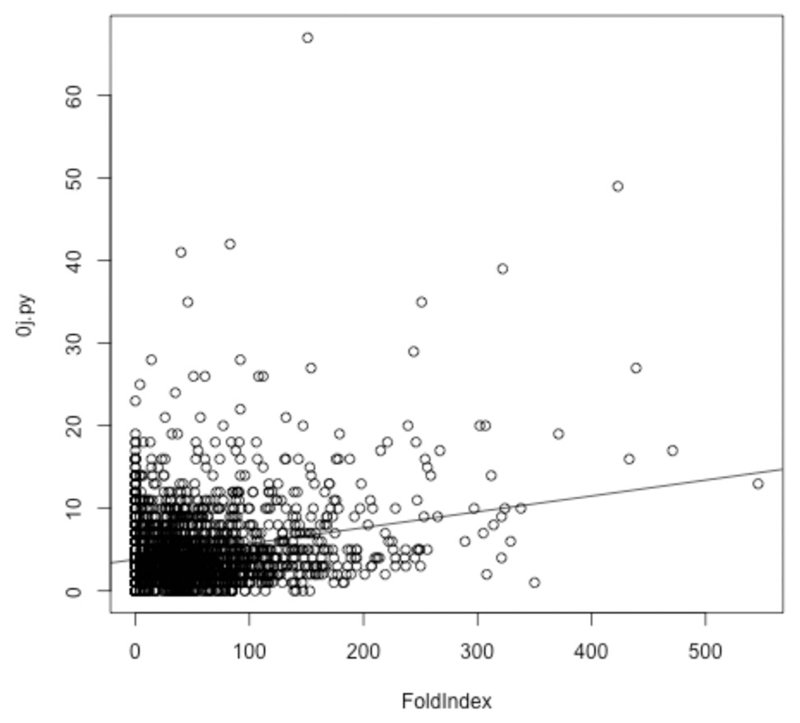
Fig. 5.
For each protein in D. radiodurans, the score reported by 0j.py, reflecting low complexity, is plotted against the number of amino acids from that protein predicted by Foldindex to be in an intrinsically disordered domain. A linear model is shown, but is a poor fit given the lack of a correlation between low complexity (as measured by 0j.py) and intrinsic disorder. Use of SEG produces very similar results (data not shown).