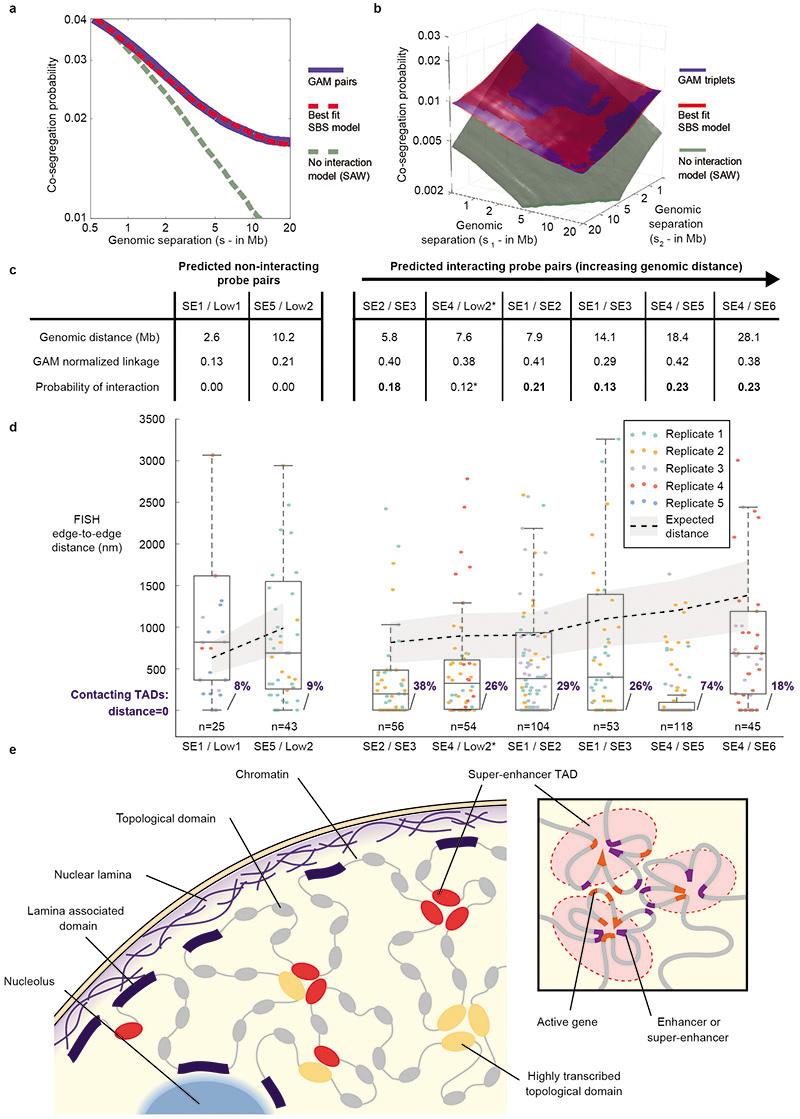
Extended Data Figure 11. Model for chromatin organization in mESC nuclei.
a and b, Polymer modelling performed using the Strings & Binders Switch (SBS) model under different conditions. We sampled the ensemble of (1) polymers in the coil thermodynamics state, equivalent to the random-open conformation of a Self-Avoiding Walk (SAW) model; (2) polymers in the compact state, where binder-specific interactions prevail and fold the polymer in closed conformations; and (3) mixtures of the SBS polymers in coil and compact states. From these in silico models, we calculated the co-segregation frequency of polymer bead pairs (a) or triplets (b) for a wide range of genomic lengths (from 0.5 up to 20 Mb). The long-range decay of co-segregation probability observed in the mESC-400 dataset (blue line and surface) is not consistent with the SAW model that lacks specific interactions (grey line and surface). Instead, the observed decay of pairs or triplets are closely matched by a 40:60 mixture of coil/compact SBS polymers (best-fit SBS model: red line, RSS =1%; red surface RSS =2%), consistent with pair and triplet contacts being abundant features of chromatin folding across large genomic distances. c, Comparison of GAM normalized linkage and SLICE Probability of Interaction (Pi) between tested pairs of TADs. d, Distribution of inter-TAD distances obtained from cryoFISH data. Grey shading and dashed black line give the median distance expected between non-interacting TADs at different linear separations. e, The chromatin fibre is organized in topologically associating domains (TADs). Inactive TADs often coincide with LADs and are therefore generally associated with either the nuclear lamina or the surface of the nucleoli. Highly transcribed TADs and TADs containing strong enhancers form clusters away from the nuclear periphery. Inset: contacts within and between highly transcribed or strong enhancer TADs are nucleated by active genes and enhancer elements.