Fig. 6.
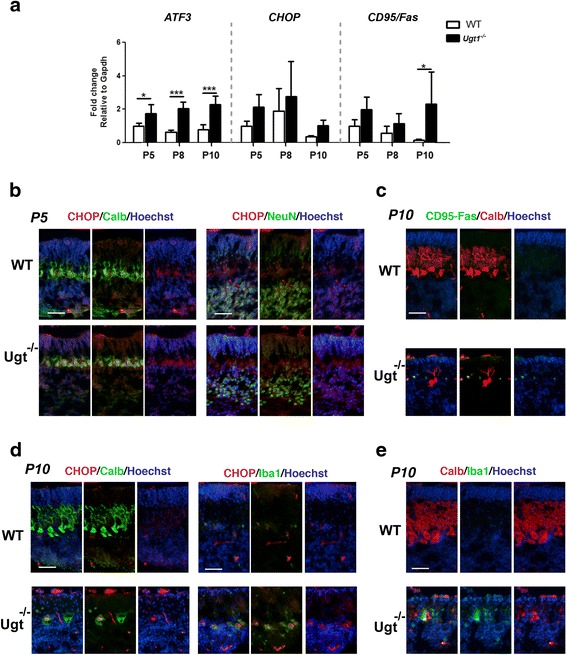
ER stress response to sustained bilirubin levels. a Relative mRNA expression analysis of different genes to assess the ER stress response over prolonged bilirubin exposure. WT and Ugt1 -/- mice mRNA expression levels of ATF3, CHOP and CD95/Fas were analysed at indicated time points by qRT-PCR. For each gene, data were normalized according to the values of the WT samples at P5. For all the experiments values represent the mean ± SD. Two-way ANOVA test, *p < 0.05. The number of WT and Ugt1 -/- was ≥3 in all the experiments and time points. b Representative IF of cerebellar sections from WT and Ugt1 -/- mice at P5 using an anti-CHOP antibody (red), co-stained with (left panel) an anti-calbindin antibody (green) to highlight Purkinje cells or (right panel) anti-NeuN to highlight granule cells. c Representative IF of cerebellar sections at P10 using an anti-CD95/Fas antibody (green), an anti-calbindin antibody (red) to highlight Purkinje cells. d Representative IF of cerebellar sections at P10 using an anti-CHOP antibody (red), co-stained with (left panel) an anti-calbindin antibody (green) or (right panel) anti-Iba1 to highlight microglia. e Representative IF of cerebellar sections at P10 using an anti-Calbindin antibody (red), co-stained with an anti-Iba1 antibody (green). For IF, Hoechst dye (blue) was used to mark nuclei. Scale bar 50 μm
