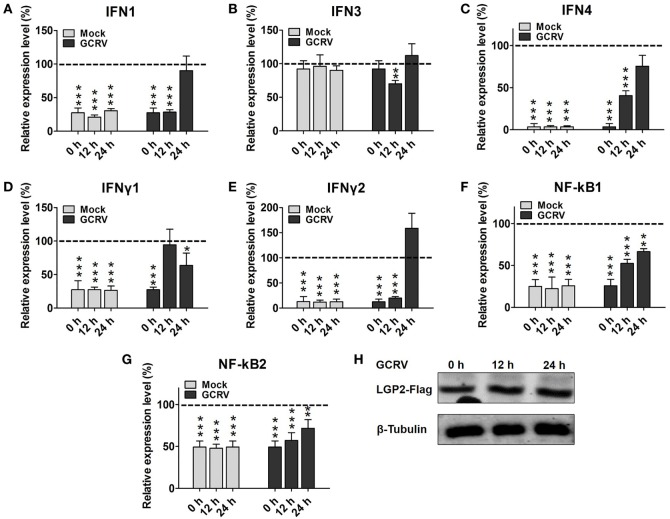
Figure 2.
Laboratory of genetics and physiology 2 (LGP2) overexpression decreases grass carp reovirus (GCRV)-induced IFNs and NF-κBs transcriptions at early stage. LGP2-Flag and empty vector (pCMV-pCMV-GFP) stable transfected Ctenopharyngodon idella kidney (CIK) cells were seeded in 12-well plates with about 70–90% monolayer confluency. Twenty-four hours later, the cells were infected with GCRV for 12 and 24 h or time-matched mock treatment. Then the cells were harvested for qRT-PCR to quantify the relative expression levels of IFN1 (A), IFN3 (B), IFN4 (C), IFNγ1 (D), IFNγ2 (E), NF-κB1 (F), and NF-κB2 (G), respectively. Fold change was determined relative to corresponding treatment group in empty vector (dash line). Error bars indicate significant differences from control (*0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001). (H) Analysis of LGP2-Flag post GCRV infection. LGP2-Flag stable transfected CIK cells were seeded in 6-well plates. Then the cells were treated with or without GCRV for 12 and 24 h. The cell lysate was used for WB analysis with anti-Flag and anti-β-Tubulin Abs.