FIGURE 4.
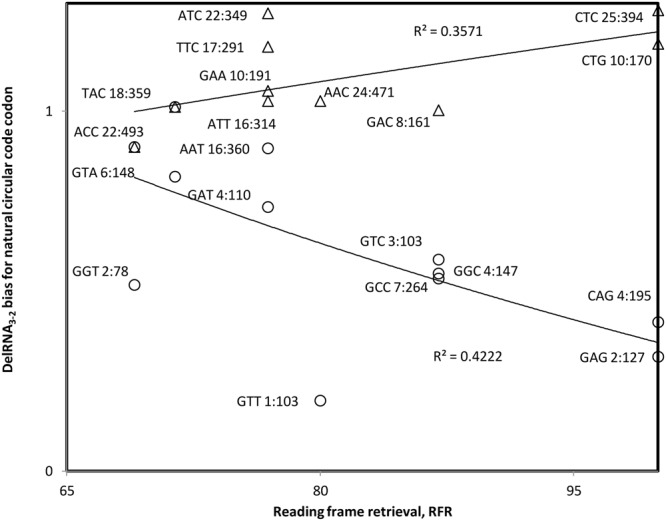
Bias in natural circular code codon contents in detected delRNA3-2 as compared to the rest of the human mitogenome, as a function of the contribution of that codon to reading frame retrieval, RFR. Natural circular code codon identities are indicated near datapoints, together with numbers of codons within delRNAs, followed by that number in the rest of the del-transformed human mitogenome. Considering all datapoints, the trend indicates avoidance of codons with high RFR (r = –0.197, P = 0.203; rs = –0.097, P = 0.342, one tailed tests). Trends seems opposite for codons with bias above versus below ‘1’, suggesting translational frame stabilizing constraints for codons with bias > ‘1’ (r = 0.598, P = 0.068; rs = 0.532, P = 0.114, two tailed tests), and transcriptional frameshifting constraints for bias < ‘1’ (r = –0.65, P = 0.011; rs = –0.606, P = 0.019, one tailed tests).
