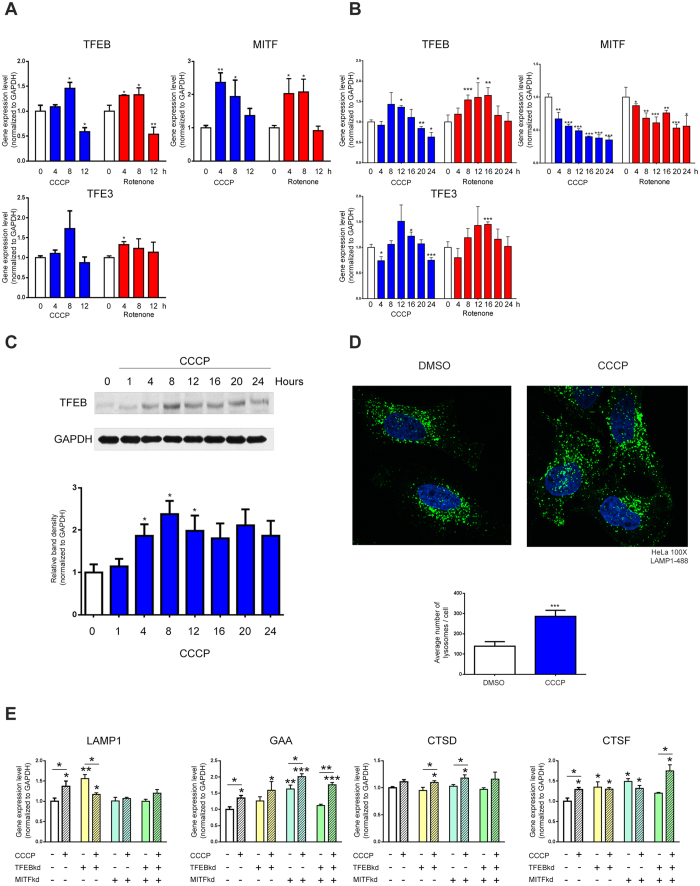
Figure 3. Acute mitochondrial stress triggers TFEB/MITF-dependent lysosomal biogenesis.
(A) Relative transcript levels of TFEB, MITF and TFE3 in MEFs treated with DMSO (controls, white bars), rotenone (250 nM, red) and CCCP (10 μM, blue), collected at 0, 4, 8 and 12 hours of treatment, normalized to GAPDH mRNA transcript level. The bars represent average and S.E.M. an (N = 4) with p-values indicated (*p < 0.05; **p < 0.005; ***p < 0.001). (B) Relative transcript levels of TFEB, MITF and TFE3 in HeLa cells treated with DMSO (controls, white bars) rotenone (250 nM, red) and CCCP (10 μM, blue), collected at 0, 4, 8, 12, 16, 20 and 24 hours of treatment, normalized to GAPDH mRNA transcript level. The bars represent average and S.E.M. (N = 6) with p-values indicated (*p < 0.05; **p < 0.005; ***p < 0.001). (C) Western blot analysis of whole-cells extracts for TFEB in HeLa cells treated with CCCP (10 μM), collected at 0, 1, 4, 8, 12, 16, 20 and 24 hours of treatment, and corresponding quantification (normalized to GAPDH). The white bar corresponds to the control HeLa cells and the blue bars to the HeLa cells treated with CCCP. The bars represent average and S.E.M. (N > 4) with p-values indicated (*p < 0.05; **p < 0.005; ***p < 0.001). (D) Representative spinning-disk microscopy images of HeLa cells treated 4 hours with CCCP 10 μM. The cells were immunostained with LAMP1-488 and DAPI. The number of the lysosomes was quantified using ImageJ. The average number of lysosomes and respective standard error of the mean are represented in the graph below. At least 15 cells of each group were used for the quantification. The statistics was done using t test, and the p-value is indicated (*p < 0.05; **p < 0.005; ***p < 0.001). (E) Transcript levels of LAMP1, GAA, CTSD and CTSF mRNA transcript levels in TFEB/MITFkd HeLa cells treated 4 hours with DMSO or CCCP (10 μM) normalized to GAPDH. The bars represent average and S.E.M. (N = 4) with t test p-values indicated (*p < 0.05; **p < 0.005; ***p < 0.001).