Figure 2.
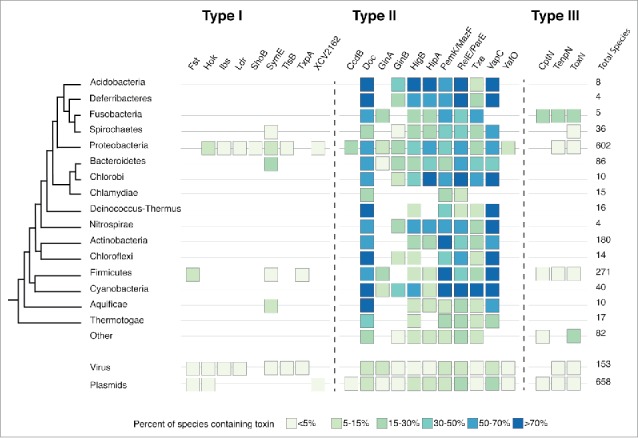
Percent of species within each phyla or replicon that contain a loci from a given type I, type II and type III TA toxin family. HMMs for each TA family were derived from known amino acid sequences and used to search a database of phage, plasmid, and bacterial chromosome sequences subjected to 6-frame translations to derive all possible amino acid sequences from that sequence. This includes short ORFs that are typical of type I toxins. For each phyla or replicon, the percent of total species in the database (left of figure) that contain at least one locus of that toxin is reported (boxes).
