Fig. 2.
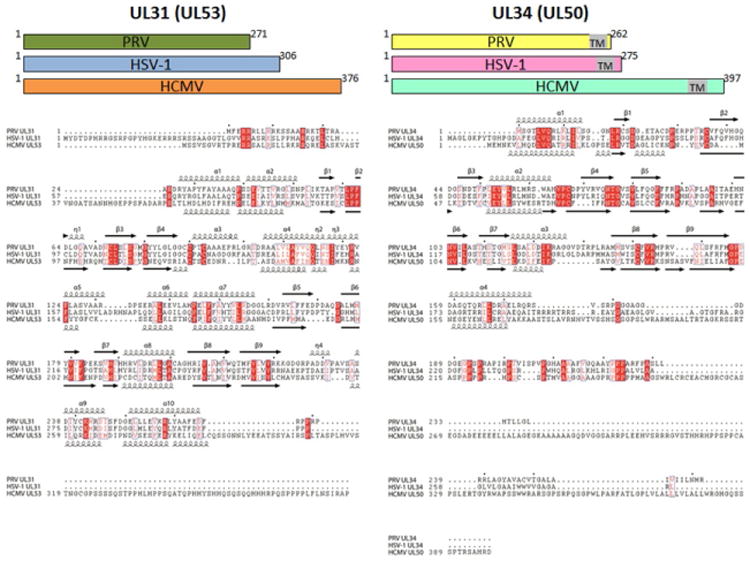
Sequence alignment of UL31 and UL34 proteins from PRV, HSV-1, and HCMV. Secondary structure for PRV and HSV-1 proteins is indicated above the sequences and for HCMV proteins, below the sequence alignment. HCMV bears additional residues at the C-termini of UL31 and UL34, which were absent from the crystallized constructs. Sequence alignment was done with Clustal Omega (Sievers and Higgins, 2014) and visualized with ESPript 3 (Robert and Gouet, 2014).
