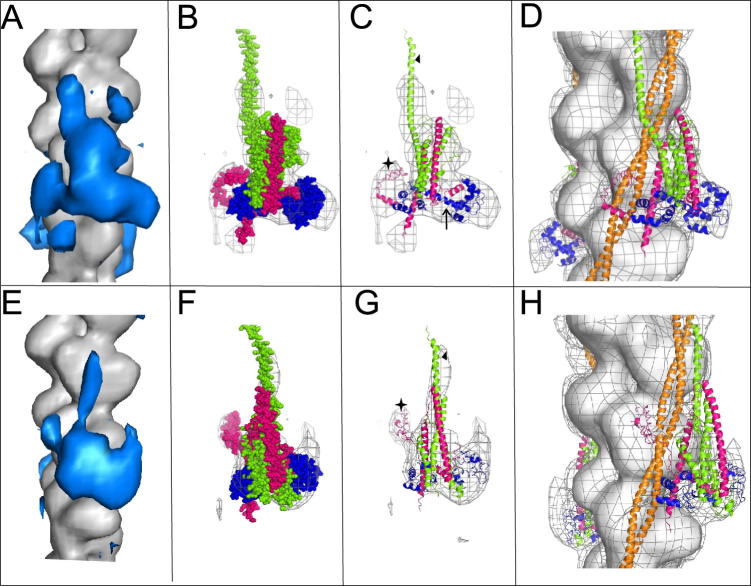
Fig. 3.
Fitting the troponin density. (A–D) Data from Ca2+-treated filaments, (E–H) data from Ca2+-free filaments. (A and E) The density attributable to troponin is shown (blue) in the Ca2+-treated state (A) and Ca2+-free state (E). (B, C and F, G) Space filling (B and F) and ribbon diagrams (C and G) of models of the troponin complex based on the crystal structures of the core domains: 1YVO for the Ca2+-free state and 1YVTZ for the Ca2+-treated state (Vinogradova et al., 2005) together with the atomistic model of the whole complex (Manning et al., 2011) that returned the best fit docked into the electron density envelopes produced by the difference analysis. The three troponin components are displayed in different colours: TnC, blue; TnT green; TnI pink. (C and G) The location of the C-terminal region of TnI is highlighted by a star, TnT1 is indicated by a triangle and the ordered central helix of TnC is arrowed in C. (D and H) Maps of the Ca2+-treated (D) and Ca2+-free (H) filaments with all the components docked into the density (tropomyosin shown in orange).