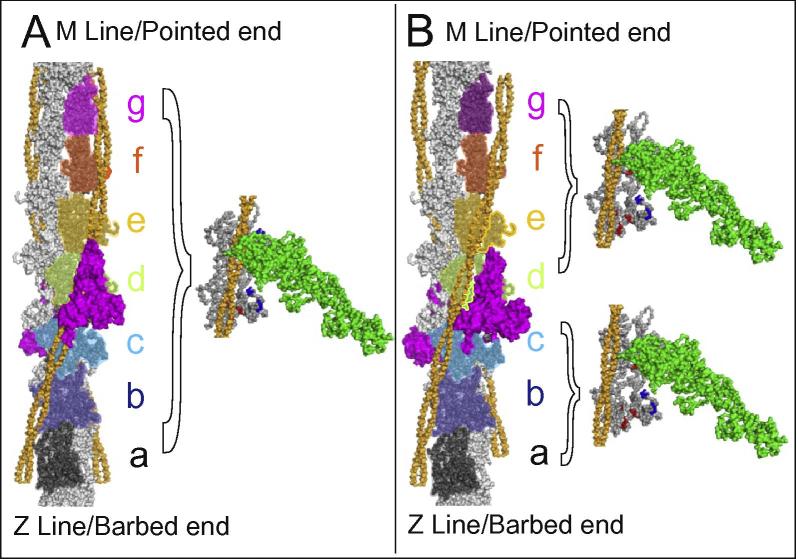
Fig. 4.
Visualisation of the tropomyosin domain movements and myosin binding. (A) Structures of the Ca2+-free state and (B) structures of the Ca2+-treated thin filament assembly. (A and B) show the central region of the Ca2+-free (A) and the Ca2+-treated (B) reconstructions over a length of 14 actin subunits, two strands of tropomyosin (orange) and two troponin complexes. The core domains of the COMPHI model are in magenta. Subunits a to g of one strand of actin are colour coded and the second strand is grey. In the Ca2+-free state (A) the position of tropomyosin is the same on every subunit and lies in the B or blocked position where it would inhibit myosin binding. The position of tropomyosin is different on different subunits of the actin filament in the Ca2+-treated state (B), where the average positions are illustrated for actin subunits d to g (closed state) and a to c (M-state). A & B (right inserts) show face on views of the actin subunits, with weak (blue) and strong (red) actin binding sites highlighted for the three distinct positions of tropomyosin. Subunits a, b and c show tropomyosin in the “M or Myosin state”, subunits d, e, f, g show tropomyosin in a position more closely aligned to the “C or closed” state. In the Ca2+-treated thin filament a myosin S1 head (green) can access all weak and potentially some strong binding sites on subunits e, f and g and presumably all available binding sites on subunits a and b. An equivalent myosin head would be blocked on every subunit in the Ca2+-free state.