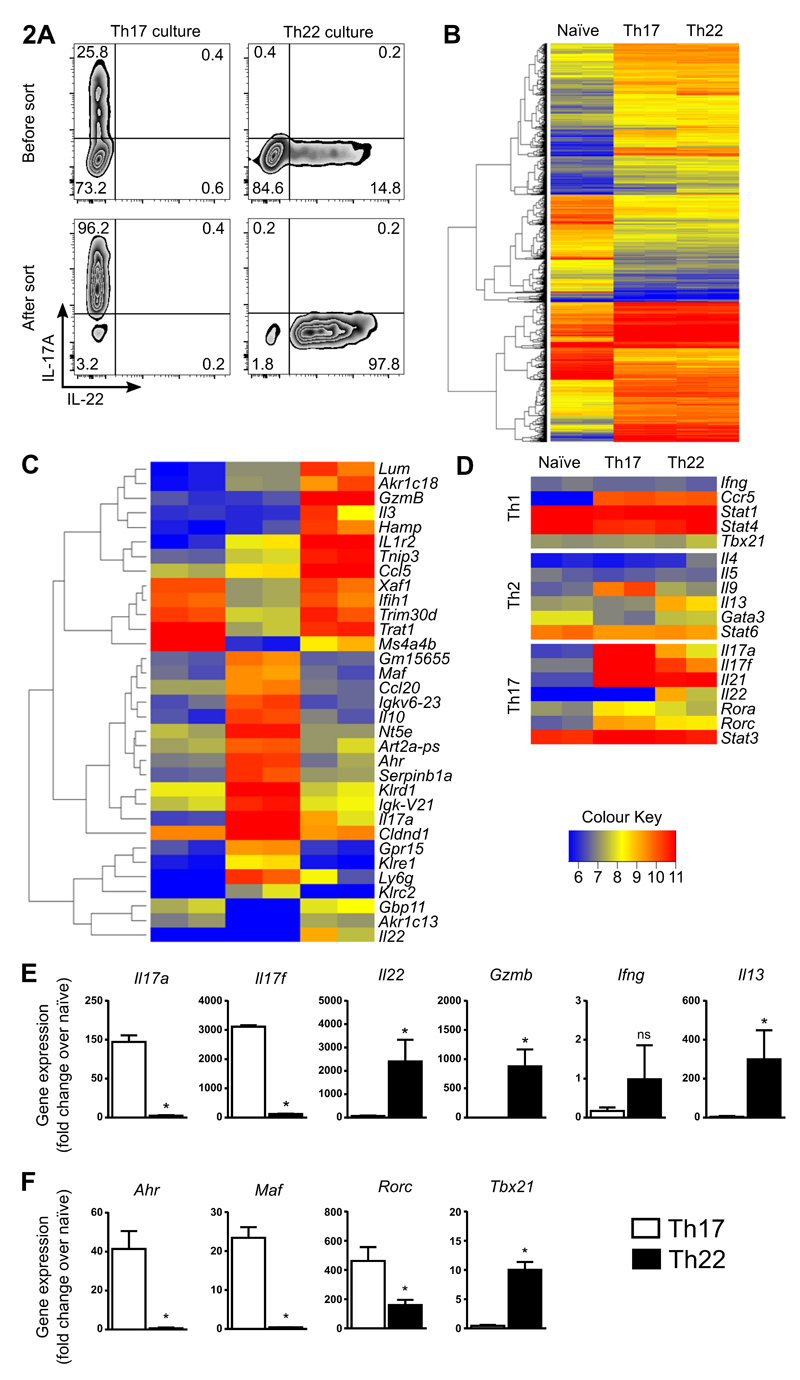
Figure 2. Transcription profiling in sorted pure Th17 and Th22 cells.
Cultures of enriched Th17 and Th22 cells were generated from IL-17eGFP x IL-22tdTomato reporter mice using optimal polarizing conditions for the first 3 d, and cells sorted on day 4 for Th17 cells (CD4+CD44+IL-17eGFP+) or Th22 cells (CD4+CD44+IL-17eGFP-IL-22tdTomato+). RNA was extracted from sorted cells and mRNA analysis performed. Th17 and Th22 enriched cultures before FACS, and resulting populations after cell sorting (viable cells shown) (A). Overview heatmap showing differentially expressed mRNAs in sorted naïve Th, Th17 and Th22 cells (B). Heatmap with 33 differentially regulated genes between Th22 and Th17 cells (>6-fold) shown for naïve Th, Th17 and Th22 cells (C). Th cell signature genes (Th1, Th2 and Th17) shown for naïve Th, Th17 and Th22 cells (D). RT-PCR confirmation of cytokine expression (E) and transcription factors (F) in sorted Th17 and Th22 cells (expression is normalized to naïve T cells). Error bars represent SEM (n=6 per group from three independent experiments). *p < 0.05.