Abstract
BACKGROUND: Irinotecan (IRI) is an integral part of colorectal cancer (CRC) therapy, but response rates are unsatisfactory and resistance mechanisms are still insufficiently understood. As fibroblast growth factor receptor 3 (FGFR3) mediates essential survival signals in CRC, it is a candidate gene for causing intrinsic resistance to IRI. METHODS: We have used cell line models overexpressing FGFR3 to study the receptor's impact on IRI response. For pathway blockade, a dominant-negative receptor mutant and a small molecule kinase inhibitor were employed. RESULTS: IRI exposure induced expression of FGFR3 as well as its ligands FGF8 and FGF18 both in cell cultures and in xenograft tumors. As overexpression of FGFR3 mitigated IRI-induced apoptosis in CRC cell models, this suggests that the drug itself activated a survival response. On the cellular level, the antiapoptotic protein bcl-xl was upregulated and caspase 3 activation was inhibited. Targeting FGFR3 signaling using a dominant-negative receptor mutant sensitized cells for IRI. In addition, the FGFR inhibitor PD173074 acted synergistically with the chemotherapeutic drug and significantly enhanced IRI-induced caspase 3 activity in vitro. In vivo, PD173074 strongly inhibited growth of IRI-treated tumors. CONCLUSION: Together, our results indicate that targeting FGFR3 can be a promising strategy to enhance IRI response in CRC patients.
Introduction
Irinotecan (IRI) is a topoisomerase inhibitor causing DNA damage and apoptosis [1]. It is used as a standard chemotherapy drug for colorectal cancer (CRC) usually in combination with 5-fluorouracil and leucovorin. However, with only 56%, response rates are unsatisfactory [2]. Mechanisms of resistance have been intensely studied, mainly focusing on pharmacokinetic parameters. Drug metabolism and uptake, carboxylesterases, and ATP-dependent drug efflux pumps have been shown to impact on individual drug response (reviewed in [3]). Much less is known about the role of DNA-damage response and survival signaling in IRI resistance [3], [4].
Fibroblast growth factor (FGF) signaling plays a central role in the protection of tissues against toxic damage [5]. As it is dysregulated in many tumor types [6], [7], it may well be a major contributor to the mitigation of drug response. In CRC, FGF18 is upregulated in a wnt-dependent manner [8], [9] and mediates tumor cell survival via the FGF receptor FGFR3-IIIc [10], [11]. FGFR3 plays a role in developing intestinal mucosa, mediating growth and morphogenesis signals induced by FGF9 and FGF18 [12]. In colonic adenomas, both FGF18 and FGFR3-IIIc are upregulated in the CD44-positive stem-like cell fraction, where they act as strong mediators of cell growth and survival [13]. Finally, whereas FGFR3 is downregulated in CRC, its IIIc splice variant is retained or even upregulated in advanced tumors [10]. Blockade of FGFR3-IIIc has been shown to inhibit colony formation, induce apoptosis in CRC cell lines, and block tumor growth in vivo [10].
Based on the FGFR3-mediated survival activity in CRC cells, the receptor has the capacity of interfering with therapy response. Therefore, the present paper investigates the role of FGFR3 overexpression in CRC response to IRI by using receptor overexpressing cell line models and FGFR3 pathway blockade to investigate options of combination therapy.
Methods and Materials
Cell Culture
SW480, SW620, HCT116, and DLD-1 cells were maintained in minimal essential medium containing 10% fetal calf serum (GE Healthcare) and passaged twice a week at 80% to 90% confluence. Caco-2 cells were maintained in minimal essential medium with 20% fetal calf serum and passaged once a week. All cell lines were obtained from the ATCC and authenticated by Eurofins Genomics (Vienna, Austria).
Overexpression of FGFR3
pcDNA constructs designed for overexpression of FGFR3-IIIb and FGFR3-IIIc were a generous gift from D. J. Donoghue. Allele-specific FGFR4 constructs FGFR4arg and FGFR4gly of the polymorphism G388R were kindly provided by A. Ullrich. Transfected SW480 cell lines overexpressing FGFR3 splice variants (FGFR3bhi or FGFR3chi) and FGFR4 polymorphic variants (FGFR4arg or FGFR4gly) were available from previous studies [10], [14]. SW620 cells overexpressing FGFR3 were generated as described previously [10].
Compounds
Oxaliplatin and IRI were from Fresenius Kabi (Graz, Austria). SN-38 was kindly provided by R. Mader (General Hospital Vienna, Austria). PD173074 was obtained from Sigma (P2499, St. Louis, MO).
Pathway Blockade
For specific inhibition of FGFR3 signaling, cells were transduced with 10 multiplicities of infection of an adenoviral vector expressing a kinase-dead mutant FGFR3-IIIc (adKD3) [9] or the control virus (adCo). Virus was added to the cultures 4 hours after exposure to IRI, and cell viability was assessed after a total incubation time of 72 hours.
Cell Growth Assay
Cells were seeded in 96-well-plates with 5 × 103/well for testing individual compounds or 2 × 103/well for combination experiments. They were treated with different drug concentrations for 72 hours. Cultures were fixed with 10% TCA for 1 hour and then stained with sulforhodamine B (SRB, Sigma) for 15 minutes and washed with 1% acetic acid. The stain was dissolved with 10 mM TRIS base (unbuffered, pH = 10), and absorption was measured with a plate reader (Tecan infinite m200, Männedorf, Switzerland) at 570 nm.
IC50 values were calculated using the nonlinear regression model in GraphPad Prism software. Combination treatments were analyzed by calculation of combination indices using CompuSyn software. Additive area was selected as 0.9 to 1.1 and above/below for antagonism/synergy, respectively [15], [16].
Cell Cycle Distribution
Cells were seeded in 6-cm dishes and exposed to the test compounds. Nuclei were isolated and stained with PI as described before [17], [18]. Samples were measured with FACSCalibur (BD).
Determination of Apoptosis
Cells undergoing apoptosis were quantified by FACS analysis after staining with the mitochondrial tracking dye JC-1 that detects loss of mitochondrial membrane potential (Sigma). Assessment of caspase-3 activity was performed according to the protocol of Werner et al. [19]. Shortly, protein was isolated from cells by homogenization in lysate buffer, and enzyme activity was measured by incubation with the specific fluorescent caspase substrate (sc-311,274, Santa Cruz Biotech) for 90 minutes. Substrate diluted in lysis buffer served as a negative control. Fluorescence was measured with a TECAN Infinite M200 plate reader.
Quantification of Gene Expression
RNA was isolated from 70% to 80% confluent plates, and 2.5 μg RNA was used for cDNA synthesis. TaqMan probes (Life Technologies, Carlsbad, CA) were used to detect FGFR1; R2; R3-IIIb; R3-IIIc; R4; and FGF8, 9, 18 (Supplementary Table 1). Average Ct values were normalized to GAPDH, and fold gene expression to untreated was plotted using the 2−∆∆Ct method [9].
Western Blot
Cells were homogenized in lysis buffer [14] using liquid nitrogen after treatment. Twenty micrograms of protein was used for Western blotting on PVDF membranes (GE Healthcare). Antibodies used were phospho H2AX [Ser138, #2577, Cell Signaling Technology (CST), Danvers, MA], bcl-xl (sc-8392, Santa Cruz Biotech), cyclin-D1 (#2926, CST), cyclin b1 (#4138, CST), phospho-cdk1 (Y15, #9111, CST), FGFR3 (#4574, CST), and beta-actin (A5441, Sigma-Aldrich).
Animals
Severe combined immunodeficient CB-17scid/scid mice (male, age 8 to 10 weeks; Harlan Laboratories, San Pietro al Natisone, Italy). The animals were kept in a pathogen-free environment, and every procedure was done in a laminar airflow cabinet. The experiments were done according to the regulations of the Ethics Committee for the Care and Use of Laboratory Animals at the Medical University Vienna (proposal number BMWF-66.009/0084-II/3b/2013), the US Public Health Service Policy on Human Care and Use of Laboratory Animals, as well as the United Kingdom Coordinating Committee on Cancer Prevention Research's Guidelines for the Welfare of Animals in Experimental Neoplasia.
Experimental Combination Therapy In Vivo
For treatment experiments, mice were inoculated with 106 HCT116 cells in 50 μl of serum-free medium subcutaneously into the right flank. When the tumors were palpable, therapy was initiated: 4 mg/kg IRI (dissolved in 50 μl of 5% dextrose) was injected i.p. twice a week in week 1 and 3 (days 13, 16, 27, and 30), and 20 mg/kg PD173074 in the same solvent was given i.p. four times in week 2 (days 21-24). Animals in the control group received solvent only. Animals were controlled for distress development every day, and tumor size was assessed regularly by caliper measurements. Tumor volume was calculated using the following formula: (length × width2)/2. The experiment was terminated at the end of week 3, and tumors were isolated and snap frozen for RNA and protein isolation.
Statistics
Statistical analysis was performed using Student's t test, analysis of variance (ANOVA), or two-way ANOVA as appropriate, and significance was designated as * for P < .05, ** for P < .01, *** for P < .001, and **** for P < .0001. Error bars depict ±SD for n ≥ 3.
Results
FGFR3-Induced Resistance to IRI
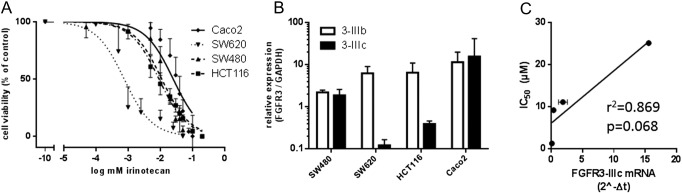
Caco2 and HCT116 cells were used as CRC models expressing high levels of FGFR3-IIIc, whereas SW480 and SW620 CRC cell lines expressed low levels of the receptor [10]. They were exposed to IRI, and IC50 concentrations were calculated. In addition, the expression of FGFR3-IIIb and FGFR3-IIIc was determined (Figure 1A; Supplementary Table 1). Sensitivity to IRI was 1.3 μM for SW620 and around 10 μM for SW480 and HCT116 cells. Caco-2 cells were the least sensitive cell line with an IC50 of 25 μM. FGFR3 expression was moderate to low in the cancer cell lines except for Caco2, which expressed the highest levels of both receptor isoforms. The lowest expression for FGFR3-IIIb was observed in SW480, and the lowest level of FGFR3-IIIc was seen in SW620 (Figure 1B, Supplementary Table 1). Although expression of FGFR3-IIIb was not related to IRI sensitivity, a rough, however not significant, correlation was found with the level of FGFR3-IIIc (Figure 1C).
Figure 1.
IRI dose response curve for standard cell lines.
(A) Cells were exposed to IRI for 72 hours, and viability was determined by SRB assay. The values represent the pooled data from three independent experiments (mean ± SD). The IC50 values and 95% CI given in the main text were calculated from these viability data using a nonlinear regression model in GraphPad Prism.
(B) RNA was isolated from semiconfluent cultures, and the baseline expression of the FGFR3 splice variants IIIb and IIIc was determined by qRT-PCR. The results confirm published data [10].
(C) The correlation between FGFR3-IIIc mRNA levels and IC50 towards IRI was calculated using GraphPad Prism software.
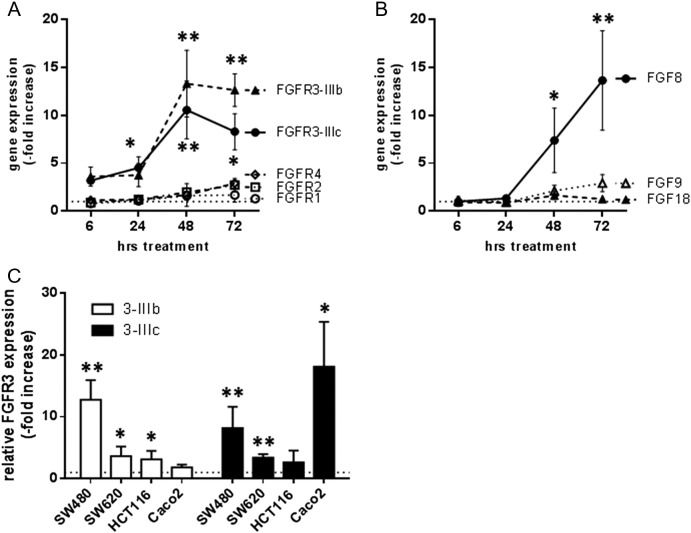
To determine the response of the FGFR signaling axis to IRI, we exposed SW480 cultures to 10 μM IRI and isolated RNA after 6 to 72 hours. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was used to determine expression of the FGFs 8, 9, and 18 as well as FGFRs 1, 2, 3b, 3c, and 4. We observed strong upregulation of both isoforms of FGFR3 in the IRI-treated groups, which reached 10-fold induction after 48 hours (Figure 2A). Other FGFRs were only little affected. In addition, the FGFR3 ligand FGF8 was similarly induced, whereas FGF18 that is highly expressed from the start [10] was not further enhanced (Figure 2B). To determine whether other cell lines upregulated the FGFR3 axis in a similar manner, cultures were exposed to IC50 concentrations of IRI, and FGFR3 expression was analyzed 72 hours later. Significantly increased mRNA levels of at least one splice variant were found in all cell lines of our panel (Figure 2C).
Figure 2.
IRI-induced expression of FGFs and FGFRs
(A, B) SW480 cells were exposed to 10 μM IRI, and RNA was isolated after the indicated intervals. RNA levels of the FGFRs 1, 2,3b, 3c, and 4 (A) and of the FGFs 8, 9 and 18 (B) were determined by qRT-PCR using Taqman assays [10]. Expression levels are given relative to the housekeeping gene GAPDH.
(C) Cultures were exposed to IC50 concentrations of IRI for 24 hours before isolation of RNA. Expression of FGFR3 splice variants was analyzed by qRT-PCR as described above. Expression of the IRI-treated cultures is presented as fold increase above control level.
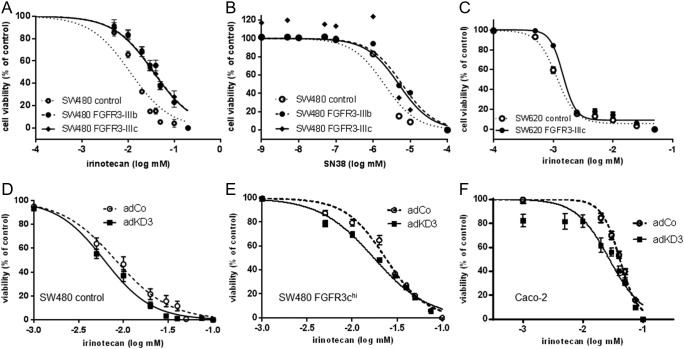
Given these results, we wondered whether FGFR3 overexpression would lead to a change in IRI sensitivity. SW480-FGFR3hi CRC cells (Supplementary Figure 1, A and B [10]) were exposed to IRI, and IC50 concentrations were calculated. We detected a significant shift to the right in the dose-response curves of SW480-FGFR3hi cells when compared with control cells (Figure 3A; P < .001 by two-way ANOVA), and IC50 concentrations increased from 11.3 μM [95% confidence interval (CI) = 9.8-12.5] in control cells to 37.2 μM (95% CI = 31.8-42.5) in FGFR3-IIIbhi cells and 36.2 μM (95% CI = 31.5-40.6) in FGFR3-IIIchi cells. As IRI is metabolically activated in the patients' liver forming SN-38 [1], dose-response to SN-38 was also assessed, and the difference of sensitivity between FGFR3hi cells and control was between 2.5- and 3-fold: 6.0 nM (95% CI = 5.5-6.7) for FGFR3-IIIb and 4.9 nM (95% CI = 3.2-7.6) for FGFR3-IIIc as compared with 2 nM (95% CI = 1.7-2.4) for control cells (Figure 3B; P < .001). SW620-FGFR3-IIIchi (Supplementary Figure 1, C–E) also displayed a significantly decreased response to IRI. The IC50 concentration was 1.5 μM (95% CI = 1.4-1.6) in FGFR3-IIIc transfectants as compared with 1.14 μM (95% CI = 1.08-1.20) in control cells (Figure 3C, P < .01).
Figure 3.
Impact of FGFR3 signaling on IRI sensitivity.
(A–C) FGFR3-overexpressing SW480 (A, B) and SW620 (C) cells were exposed to IRI (A, C) or SN38 (B) for 72 hours. Cell viability was measured by SRB assay, and dose-response curves were constructed by Graphpad Prism software. Values shown are mean ± SD.
(D–F) SW480 controls (D) and SW480 FGFR3hi (E) clone pools as well as Caco2 cultures (F) were exposed to increasing concentrations of IRI. Four hours later, they were transduced with an adenoviral vector expressing the dominant-negative FGFR3 mutant KD3 (AdKD3) or a control virus (AdCo). After a total incubation time of 72 hours, viability was determined and dose-response curves were constructed.
Targeting FGFR Signaling to Increase IRI Response
The highly FGFR3 expressing cell lines Caco-2 and SW480-FGFR3hi were used to study the impact of FGFR3 blockade on IRI response. For genetic blockade, we used the kinase-dead FGFR3-IIIc specific adenoviral construct adKD3 [10] that was applied 4 hours after IRI addition to prevent activity of the IRI-induced FGFR3. After 72 hours of incubation, cell viability was assessed and dose-response curves were constructed. An increase in IRI sensitivity was observed even in the SW480 control cells, and IC50 concentrations decreased from 6.8 μM (95% CI = 5.9-7.9) in the adCo group to 4.7 μM (95% CI = 4.2-5.3) in the adKD3 group (Figure 3D). With P = .051, the difference just missed statistical significance. In SW480 FGFR3chi cells, a partial reversal of the FGFR impact on IRI response was seen, and IC50 was 15.3 μM (95% CI = 14.1-16.5) in the adKD3 infected cultures as compared with 19.5 μM (95% CI = 17.6-21.5; P = .015) in the control group (Figure 3E). An even greater impact was observed for Caco-2 cells, which express high endogenous FGFR3 levels: IC50 values decreased from 45.5 μM (95% CI = 39.1-53.0) to 25.7 μM (95% CI = 21.9-30.2; P < .01) (Figure 3F).
For pharmacological blockade of the FGFR3 impact, the small molecule FGFR inhibitor PD173074 was used together with IRI. PD173074 exposure induced cell loss in all cell lines tested, and IC50 concentrations are given in Supplementary Table 2. To determine the combined impact of both drugs, SW480-FGFR3hi, SW480 control cells, SW620 transfectants, HCT116, and Caco2 cells were exposed to the single drugs as well as all possible combinations of both. Cell viability was measured after 72 hours, and dose-response curves were constructed (Supplementary Figure 3).
All viability results were analyzed by using Compusyn software to calculate combination indices. Synergistic effects at low concentrations of IRI and PD173074 were observed in all cell lines (Table 1).
Table 1.
Synergistic Effect of IRI and PD173074
| Cell Line | Combination Index⁎ | 95% CI | Used IRI/PD doses (μM) |
|---|---|---|---|
| SW480 control | 0.69 | 0.55-0.83 | 2.5/1.0 |
| SW480 FGFR3bhi | 0.73 | 0.51-0.96 | |
| SW480 FGFR3chi | 0.66 | 0.54-0.79 | |
| SW620 control | 0.87 | 0.71-1.03 | 1.25/1.0 |
| SW620 FGFR3chi | 0.71 | 0.58-0.83 | |
| Caco-2 | 0.55 | 0.13-0.97 | 10.0/1.0 |
| 0.43 | 0.02-0.88 | 10.0/5.0 | |
| HCT116 | 0.87 | 0.65-1.09 | 2.5/1.0 |
Combination Index (CI) is calculated based on the dose-response curves depicted in Supplementary Figure 2 using CompuSyn software. 0.9 < CI< 1.1 was considered to indicate an additive effect. CI< 0.9 shows synergistic interaction; CI> 1.1 means antagonistic interaction.
FGFR3 Expression Inhibits IRI-Induced Apoptosis
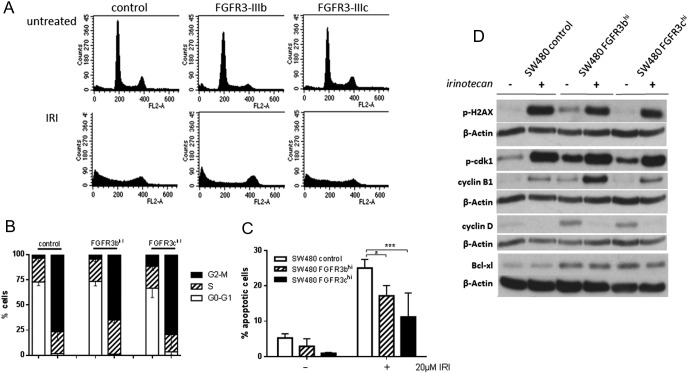
To characterize the cells' response to IRI, cultures were exposed to 20 μM IRI for 72 hours. Then cell cycle was analyzed, and protein lysates were produced to investigate DNA damage, cell cycle markers, and apoptosis induction. IRI treatment led to a strong cell cycle block in the G2/M phase, whereas G1 cells were essentially absent from the population (Figure 4, A and B). In addition, a large fraction of nuclei were registered in the sub-G1 or debris fraction, indicating massive cell death. Analysis of apoptosis using JC-1 to measure mitochondrial membrane potential demonstrated strong induction of apoptosis in SW480 cells exposed to IRI as described above. This response was significantly weaker in SW480 FGFR3hi cells than in control transfectants (Figure 4C).
Figure 4.
Cellular response to IRI.
SW480 control and FGFR3hi cells were exposed to 20 μM IRI for 72 hours before they were harvested for:
(A, B) FACS analysis of cell cycle as described in the Materials and Methods section. (A) A typical analysis. (B) The pooled data of three independent experiments as mean ± SD.
(C) Quantification of apoptosis using the JC1 mitochondrial tracker dye. Bars represent the mean ± SD of three independent experiments, and differences were analyzed by ANOVA.
(D) Protein analysis by Western blot using antibodies to p-H2AX, p-cdk1, cyclin B, cyclin D, and bcl-xl as described in Materials and Methods.
In the cell lysates, upregulation of phospho-H2AX, a prominent DNA damage marker [20], [21] was observed. Cyclin B and phosphorylated cdk1 were increased in IRI-exposed cells, whereas cyclin D was reduced, confirming the cell cycle arrest in G2 (Figure 3D). Both damage and cell cycle arrest were similar in SW480 control as well as SW480-FGFR3hi cells, indicating that they were independent of FGFR3.
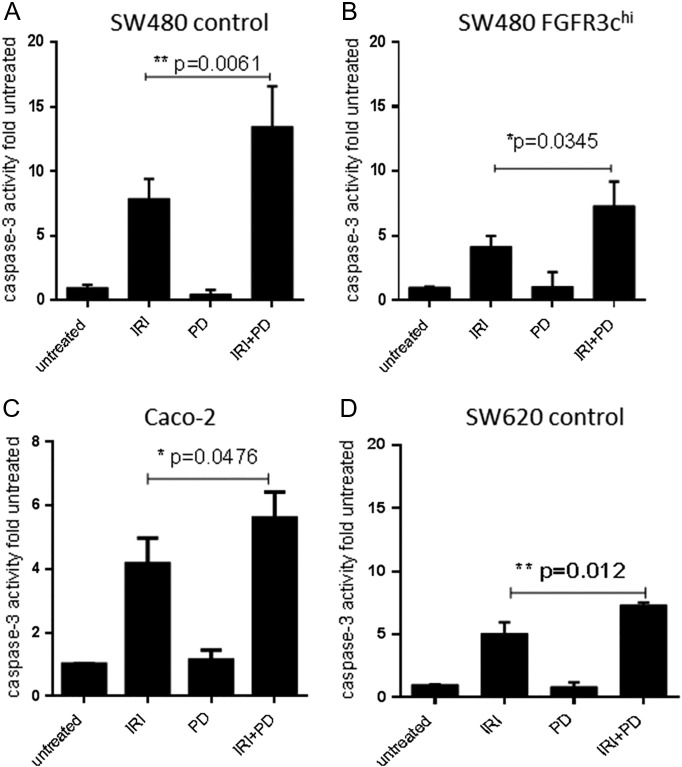
Analysis of antiapoptotic bcl-xl revealed that baseline bcl-xl protein expression was significantly upregulated in FGFR3-overexpressing cells. Whereas IRI induced a loss of bcl-xl protein in all exposed cultures, the levels remained higher in the SW480-FGFR3hi cells as compared with controls (Figure 4D; bottom panel). IRI-induced caspase-3 activity measured after exposure to synergistic concentrations of IRI and PD173074 (Table 1) was mitigated by FGFR3 overexpression (Figure 5, A and B; IRI groups). Caspase-3 was analyzed after single and combined treatment with the inhibitor PD173074 and was significantly elevated by addition of the FGFR inhibitor as compared with single treatment with IRI not only in SW480 transfectants (Figure 5, A and B) but also Caco-2 cells (Figure 5C) and even parental SW620 cells in spite of their low FGFR3 expression level (Figure 5D).
Figure 5.
IRI-induced caspase activity.
Cells were exposed to synergistic concentrations of IRI, PD173074, or both compounds for 72 hours before being lysed for determination of caspase 3 activity. Activity was calculated relative to the untreated control, and bars represent the mean ± SD of three independent experiments. Differences were analyzed by Student's t test with Welch correction.
(A) SW480 control, (B) SW480 FGFR3-IIIc, (C) Caco-2, and (D) SW620 control.
Impact on Tumor Growth In Vivo
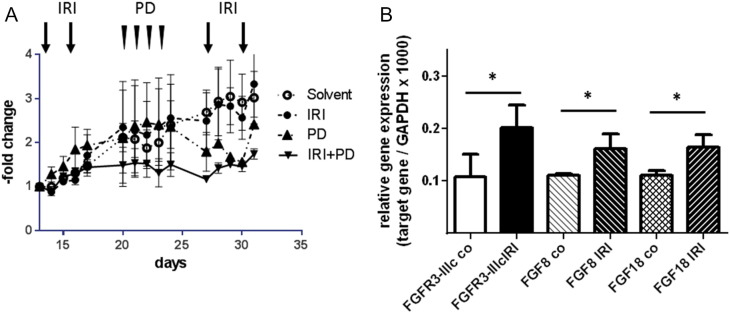
HCT116 cells were chosen to assess the efficacy of combined IRI/PD173074 treatment on tumor growth in vivo because they express high levels of FGFR3 and induce rapid tumor growth when injected subcutaneously. Cells were injected into the rear flanks of SCID mice, and treatment was started when the tumors became palpable. IRI and PD were applied sequentially: IRI on days 13 and 16 in week 1 and on days 27 and 30 in week 3; PD on days 21 to 24 in week 2. IRI alone had little impact on tumor growth, and PD alone reduced tumor size after 7 days. In the combination treatment group, PD effectively prevented further tumor growth from the start of PD treatment (Figure 6A; combination treatment different from IRI group at P = .0005 by two-way ANOVA).
Figure 6.
Combination therapy using IRI and PD173074.
SCID mice were inoculated with 106 HCT116 cells s.c. in their flank. Treatment was initiated when the tumors had become palpable and was done sequentially. Four milligrams per kilogram IRI was injected i.p. twice a week in weeks 1 and 3 (days 13, 16, 27, and 30). Twenty milligrams per kilogram PD173074 was given i.p. four times in week 2 (days 21-24). The experiment was terminated at the end of week 3, and tumors were isolated and snap frozen for the isolation of RNA.
(A) Tumor growth was monitored using a Vernier caliper until day 33 when the animals were sacrificed. It was calculated relative to tumor size on day 13 for each individual tumor, and each data point represents the mean ± SD of three tumors.
(B) RNA was isolated from the snap-frozen tumor tissue, and expression of FGFR3-IIIc, FGF8, and FGF18 was determined by qRT-PCR and calculated relative to the housekeeping gene GAPDH. Bars represent the mean ± SD of three tumors. Differences were analyzed by Student's t test with Welch correction.
Mice were sacrificed at the end of week 3 after the second series of IRI. Tumor tissue was shock frozen, and expression of FGFR3, FGF8, and FGF18 was determined by qRT-PCR. IRI-treated tumors expressed higher levels of all three genes as compared with control tumors (Figure 6B), indicating that upregulation of FGFR3 survival signaling also happens in vivo.
Discussion
Irinotecan is a topoisomerase-1 inhibitor that is used in the therapy of advanced CRC mainly in combination with 5-FU (FOLFIRI), and about 60% of the patients respond to the treatment [2]. Mechanisms of resistance have been widely studied mainly focusing on pharmacokinetic parameters — drug metabolism, uptake, and efflux pumps (reviewed in [3]). The drug needs to be activated by carboxylesterases whose expression and polymorphic alleles have impact on the effectivity of IRI [22]. Moreover, IRI is the substrate of several drug efflux pumps — specifically ABCG2 [1], ABCB1 and ABCC2 [23]. Our data now show that FGFR-dependent survival signaling may also impede IRI response. Specifically, we demonstrate that IRI upregulates FGFR3-IIIc expression, which inhibits IRI-induced apoptosis, by inducing the antiapoptotic gene bcl-xl.
Even though FGFR3 is not highly expressed in CRC, the splice variant FGFR3-IIIc mediates FGF18-dependent survival signaling in colorectal tumors from the adenoma stage [10], [13]. FGFR3-IIIc is upregulated in advanced CRC, and pathway blockade induced apoptosis [10]. Results of this study show that both endogenous expression and enforced expression of the receptor are related to low sensitivity of cells toward IRI. The IC50 concentrations are shifted by a factor of about 4 for IRI and 3 for SN38 (Figure 3). This cannot be explained by increased cell growth in the overexpressing cultures because the impact on proliferation is moderate for FGFR3-IIIc [10] (Supplementary Figure 1) and absent for FGFR3-IIIb [10]. In addition, a dominant-negative receptor mutant (KD3) increased sensitivity to IRI, and inhibition of the receptor kinase by PD173074 had a synergistic effect together with low concentrations of IRI.
FGFR3 is not the only tyrosine kinase receptor (TKR) that can interfere with the response to chemotherapy drugs as TRK downstream signaling impacts on DNA damage response [24]. Also inhibition of several TKRs has been shown to induce apoptosis [25]. Consequently, blocking antibodies targeting EGFR enhance chemotherapy response unless activating mutations in the downstream pathways (e.g., in Ki-ras) prevent pathway blockade [26], [27]. Also, small molecule inhibitors of c-met are capable of reversing resistance to IRI [28] or cisplatin [29]. Among FGFRs, the FGFR2 gene was upregulated in drug-resistant gastric cancer cell clones [30].
FGFR3 differs from these other TKRs as its expression is actually induced by exposure to the drug. This indicates that IRI-induced resistance not only acts through a long-term selection process enriching for surviving resistant clones but also in a short-term survival response that can be observed as early as 6 hours after exposure (Figure 2, A and B). This can explain why synergism between the FGFR inhibitor PD173074 and IRI was seen not only in FGFR3hi cells but in all cell models — even in SW620 cells that showed the lowest FGFR3-IIIc expression in our cell line panel. As PD173074 targets other FGFRs as well as FGFR3, we cannot exclude that these FGFRs also counteract IRI response. However, two observations argue against this possibility: 1) other FGFRs are not upregulated by exposure to the drug, and 2) FGF18, which is high in most colon cancer cells, and FGF8, which is induced by IRI, are mainly FGFR3 ligands [11]. Increased mRNA levels for FGFR3-IIIc as well as its ligands FGF8 and FGF18 were also found in xenograft tumors treated with IRI, indicating that the survival response also takes place in vivo. It may be the reason that IRI alone was not very effective against HCT116-xenograft tumors. Tumors in the IRI treatment group grew more slowly, but subsequent administration of PD173074 reduced tumor growth to almost zero. Consequently, tumors were too small to analyze efficiency of apoptosis in the tissue.
In the in vitro models, however, the impact on apoptosis induction could be clearly demonstrated. Overexpression of FGFR3 induced expression of the antiapoptotic protein bcl-xl and strongly inhibited IRI-induced caspase 3 activation. Inhibition of the FGFR3-kinase by PD173074 reversed this effect, permitting efficient induction of apoptosis. These results are well in line with our earlier reports that FGFR3 pathway blockade by KD3 alone induced apoptosis in both CRC cells in vitro and in CRC xenograft tumors [10]. FGFR3 also mediates survival in a stem-like subpopulation of cells from the early adenoma stage onward [13]. This small subpopulation should be selectively protected against IRI therapy.
Correlation of FGFR3 levels with IRI response in human CRC tissue could not be shown either from our own expression analysis or from published data sets. This is probably due to the generally low expression of the receptor in CRC tissue and to the small number of reports on IRI therapy that include gene expression data. FGFR3 expression has little promise as a predictive marker in this context. Based on the IRI-dependent induction of FGFR3 gene expression and the strong synergy seen between IRI and FGFR3 inhibition, the receptor is a promising target for combination therapy.
Acknowledgements
The study was supported by the Austrian Science Foundation (P 23693).
Footnotes
Grant support: The study was supported by the Austrian Science Foundation (P 23693).
Disclosures: None of the authors has any conflict of interest to declare.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2017.02.004.
Appendix A. Supplementary data
Supplementary material
References
- 1.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 2.Goodwin RA, Asmis TR. Overview of systemic therapy for colorectal cancer. Clin Colon Rectal Surg. 2009;22:251–256. doi: 10.1055/s-0029-1242465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panczyk M. Pharmacogenetics research on chemotherapy resistance in colorectal cancer over the last 20 years. World J Gastroenterol. 2014;20:9775–9827. doi: 10.3748/wjg.v20.i29.9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoskins JM, Marcuello E, Altes A, Marsh S, Maxwell T, Van Booven DJ, Paré L, Culverhouse R, McLeod HL, Baiget M. Irinotecan pharmacogenetics: influence of pharmacodynamic genes. Clin Cancer Res. 2008;14:1788–1796. doi: 10.1158/1078-0432.CCR-07-1472. [DOI] [PubMed] [Google Scholar]
- 5.Braun S, Auf dem Keller U, Steiling H, Werner S. Fibroblast growth factors in epithelial repair and cytoprotection. Philos Trans R Soc Lond B Biol Sci. 2004;359:753–757. doi: 10.1098/rstb.2004.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinzle C, Sutterluty H, Grusch M, Grasl-Kraupp B, Berger W, Marian B. Targeting fibroblast-growth-factor-receptor–dependent signaling for cancer therapy. Expert Opin Ther Targets. 2011 doi: 10.1517/14728222.2011.566217. [DOI] [PubMed] [Google Scholar]
- 7.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 8.Shimokawa T, Furukawa Y, Sakai M, Li M, Miwa N, Lin Y-M, Nakamura Y. Involvement of the FGF18 gene in colorectal carcinogenesis, as a novel downstream target of the {beta}-catenin/T-cell factor complex. Cancer Res. 2003;63:6116–6120. [PubMed] [Google Scholar]
- 9.Sonvilla G, Allerstorfer S, Stattner S, Karner J, Klimpfinger M, Fischer H, Grasl-Kraupp B, Holzmann K, Berger W, Wrba F. FGF18 in colorectal tumour cells: autocrine and paracrine effects. Carcinogenesis. 2008;29:15–24. doi: 10.1093/carcin/bgm202. [DOI] [PubMed] [Google Scholar]
- 10.Sonvilla G, Allerstorfer S, Heinzle C, Stattner S, Karner J, Klimpfinger M, Wrba F, Fischer H, Gauglhofer C, Spiegl-Kreinecker S. Fibroblast growth factor receptor 3-IIIc mediates colorectal cancer growth and migration. Br J Cancer. 2010;102:1145–1156. doi: 10.1038/sj.bjc.6605596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family: the complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brodrick B, Vidrich A, Porter E, Bradley L, Buzan JM, Cohn SM. Fibroblast growth factor receptor-3 (FGFR-3) regulates expression of Paneth cell lineage-specific genes in intestinal epithelial cells through both TCF4/β-catenin–dependent and –independent signaling pathways. J Biol Chem. 2011;286:18515–18525. doi: 10.1074/jbc.M111.229252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koneczny I, Schulenburg A, Hudec X, Knofler M, Holzmann K, Piazza G, Reynolds R, Valent P, Marian B. Autocrine fibroblast growth factor 18 signaling mediates Wnt-dependent stimulation of CD44-positive human colorectal adenoma cells. Mol Carcinog. 2015;54:789–799. doi: 10.1002/mc.22146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinzle C, Gsur A, Hunjadi M, Erdem Z, Gauglhofer C, Stattner S, Karner J, Klimpfinger M, Wrba F, Reti A. Differential effects of polymorphic alleles of FGF receptor 4 on colon cancer growth and metastasis. Cancer Res. 2012;72:5767–5777. doi: 10.1158/0008-5472.CAN-11-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lange J, Ly LV, Lodder K, Verlaan-de Vries M, Teunisse AF, Jager MJ, Jochemsen AG. Synergistic growth inhibition based on small-molecule p53 activation as treatment for intraocular melanoma. Oncogene. 2012;31:1105–1116. doi: 10.1038/onc.2011.309. [DOI] [PubMed] [Google Scholar]
- 16.Tsakalozou E, Eckman AM, Bae Y. Combination effects of docetaxel and doxorubicin in hormone-refractory prostate cancer cells. Biochem Res Int. 2012;2012:832059. doi: 10.1155/2012/832059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hausott B, Greger H, Marian B. Flavaglines: a group of efficient growth inhibitors block cell cycle progression and induce apoptosis in colorectal cancer cells. Int J Cancer. 2004;109:933–940. doi: 10.1002/ijc.20033. [DOI] [PubMed] [Google Scholar]
- 18.Kainz KP, Krenn L, Erdem Z, Kaehlig H, Zehl M, Bursch W, Berger W, Marian B. 2-deprenyl-rheediaxanthone B isolated from Metaxya rostrata induces active cell death in colorectal tumor cells. PLoS One. 2013;8:e65745. doi: 10.1371/journal.pone.0065745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werner M, Sacher J, Hohenegger M. Mutual amplification of apoptosis by statin-induced mitochondrial stress and doxorubicin toxicity in human rhabdomyosarcoma cells. Br J Pharmacol. 2004;143:715–724. doi: 10.1038/sj.bjp.0705928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair. 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Bellott R, Le Morvan V, Charasson V, Laurand A, Colotte M, Zanger UM, Klein K, Smith D, Bonnet J, Robert J. Functional study of the 830C>G polymorphism of the human carboxylesterase 2 gene. Cancer Chemother Pharmacol. 2008;61:481–488. doi: 10.1007/s00280-007-0493-9. [DOI] [PubMed] [Google Scholar]
- 23.Han J-Y, Lim H-S, Yoo Y-K, Shin ES, Park YH, Lee SY, Lee J-E, Lee DH, Kim HT, Lee JS. Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer. 2007;110:138–147. doi: 10.1002/cncr.22760. [DOI] [PubMed] [Google Scholar]
- 24.Chen MK, Hung MC. Regulation of therapeutic resistance in cancers by receptor tyrosine kinases. Am J Cancer Res. 2016;6:827–842. [PMC free article] [PubMed] [Google Scholar]
- 25.Zwick E, Bange J, Ullrich A. Receptor tyrosine kinases as targets for anticancer drugs. Trends Mol Med. 2002;8:17–23. doi: 10.1016/s1471-4914(01)02217-1. [DOI] [PubMed] [Google Scholar]
- 26.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 27.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 28.Yashiro M, Nishii T, Hasegawa T, Matsuzaki T, Morisaki T, Fukuoka T, Hirakawa K. A c-Met inhibitor increases the chemosensitivity of cancer stem cells to the irinotecan in gastric carcinoma. Br J Cancer. 2013;109:2619–2628. doi: 10.1038/bjc.2013.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li E, Hu Z, Sun Y, Zhou Q, Yang B, Zhang Z, Cao W. Small molecule inhibitor of c-Met (PHA665752) suppresses the growth of ovarian cancer cells and reverses cisplatin resistance. Tumour Biol. 2016;37:7843–7852. doi: 10.1007/s13277-015-4318-x. [DOI] [PubMed] [Google Scholar]
- 30.Qiu H, Yashiro M, Zhang X, Miwa A, Hirakawa K. A FGFR2 inhibitor, Ki23057, enhances the chemosensitivity of drug-resistant gastric cancer cells. Cancer Lett. 2011;307:47–52. doi: 10.1016/j.canlet.2011.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material