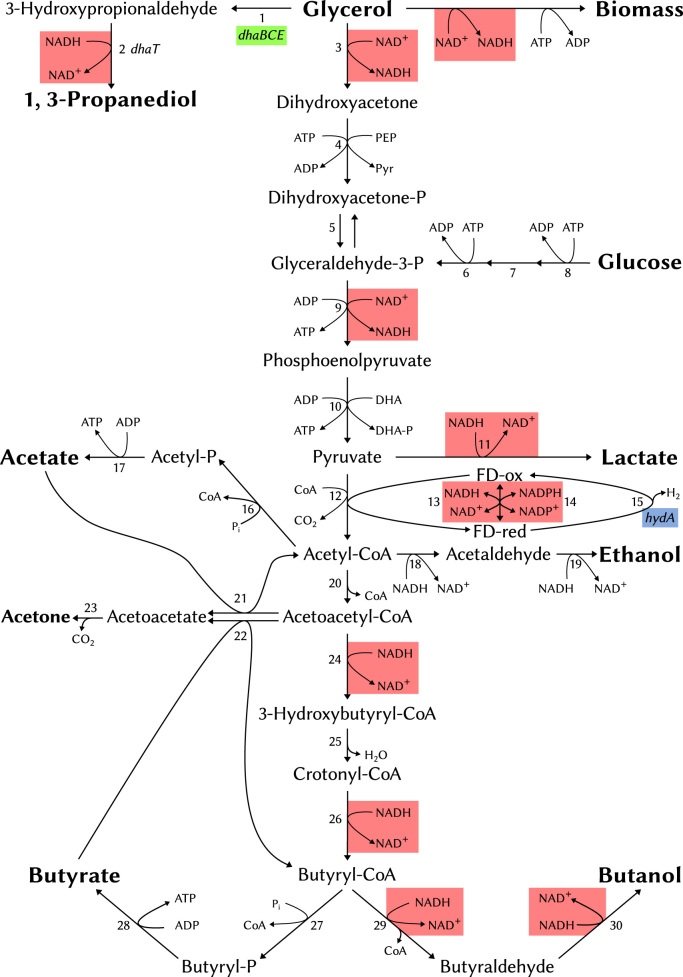
Fig. 1.
Central metabolic energy pathway in C. pasteurianum from glucose and glycerol. Figure based on Malaviya et al. (2012) and Biebl (2001). The knocked out genes (hydA, dhaBCE) and the NADH/NAD+ utilising pathways putatively influenced by Rex are highlighted. Other enzymes involved in the central energy pathway are numbered as follows: 1, glycerol dehydratase; 2, 1,3-propanediol oxydoreductase. 3, glycerol-3-phosphate dehydrogenase; 4, dihydroxyacetone kinase; 5, triose-phosphate isomerase; 6, phosphofructokinase; 7, phosphoglucose isomerase; 8, hexokinase; 9, glyceraldehyde-3-phosphate dehydrogenase; 10, pyruvate kinase; 11, lactate dehydrogenase; 12, pyruvate-ferredoxin oxidoreductase; 13, ferredoxin-NADP reductase; 14, NADPH-ferredoxin oxidoreductase; 15, ferredoxin hydrogenase; 16, phosphate acetyltransferase; 17, acetate kinase; 18, acetaldehyde dehydrogenase; 19, ethanol dehydrogenase; 20, thiolase; 21, acetoacetyl-CoA: acetate:CoA transferase; 22, acetoacetyl-CoA: butyrate:CoA transferase; 23, acetoacetate decarboxylase; 24, β -hydroxybutyryl-CoA dehydrogenase; 25, crotonase; 26, butyryl-CoA dehydrogenase; 27, phosphotransbutyrylase; 28, butyrate kinase; 29, butaraldehyde dehydrogenase; 30, butanol dehydrogenase.