Abstract
Our long-term goal is to prevent or reverse atherosclerosis by delivering gene therapy from stably transduced endothelial cells (EC). We previously reported that EC-directed gene therapy with a helper-dependent adenovirus (HDAd) expressing apolipoprotein A-I (apo A-I) retarded development of atherosclerosis in rabbit carotid arteries over a 1-month interval. However, a 70% decline in apo A-I expression during this time raised concerns about long-term efficacy of this approach. Here we report use of several approaches aimed either at preventing this decline or at increasing apo A-I expression from HDAd at all time points: codon optimization, deletion of 3′ untranslated sequences, substitution of a synthetic mammalian-based promoter (4XETE) for the cytomegalovirus (CMV) promoter, and co-transduction with an HDAd expressing interleukin-10. We tested these approaches using plasmid transfection of cultured EC and in vivo transduction of rabbit carotid artery EC. Codon optimization did not increase apo A-I expression. Deletion of 3′ untranslated sequences extinguished apo A-I expression. Both substitution of 4XETE for the CMV promoter and expression of interleukin-10 stabilized apo A-I expression in vivo, although at the cost of lower early (3-day) expression levels. Surprisingly, both interventions stabilized apo A-I expression without altering the rate at which HDAd genomes were lost. These data establish that transgene expression from HDAd in EC is inherently stable in vivo and suggest that the early decline of CMV promoter-driven expression from HDAd-transduced EC is due neither to active downregulation of transcription nor to loss of HDAd genomes. Instead, apparent loss of expression from the CMV promoter appears to be a consequence of early (3-day) upregulation of CMV promoter activity via inflammatory pathways. Our results yield new paradigms to explain the early loss of genomes and transgene expression after in vivo gene transfer. These new paradigms will redirect strategies for achieving high-level, stable expression of transgenes in EC.
Keywords: : apolipoprotein A-I, endothelial cells, gene expression, gene therapy, helper-dependent adenovirus, interleukin-10
Introduction
Vascular endothelial cells (EC) line the lumen of blood vessels and play critical roles in several important physiological processes including regulation of blood pressure and tissue perfusion, transport of macromolecules into tissues, regulation of hemostasis and thrombosis, and trafficking of leukocytes.1 Because they are in direct contact with circulating blood, EC are attractive targets for percutaneous gene delivery, for example by intravenous or intra-arterial vector injection. Moreover, because EC overlie atherosclerotic plaques and influence the biological processes that promote or prevent development of atherosclerosis,2 EC are in an ideal location to deliver gene therapy aimed at preventing or treating atherosclerosis. Accordingly, several groups have attempted to achieve stable high-level transgene expression in EC.3–6
Achievement of stable high-level transgene expression in EC requires a vector with significant endothelial tropism as well as a potent expression cassette that maintains activity indefinitely. We and others have shown that serotype 5 adenoviral vectors have high tropism for both animal and human EC.7,8 However, transgene expression in EC transduced in vivo with first- and second-generation adenoviral vectors is lost rapidly (∼2 weeks) due to a combination of immune system–mediated clearance of transduced EC and direct toxicity of viral peptides expressed from the adenoviral vector backbone.9 This limitation is overcome by use of helper-dependent adenoviral (HDAd) vectors, which express no viral proteins10 and provide long-term (at least 48 weeks) transgene expression in EC.11 However, in most cases, initial high levels of transgene expression in HDAd-transduced EC decline substantially between 3 and 28 days after transduction and then stabilize.11,12 This early decline in transgene expression could limit the long-term efficacy of EC-targeted gene therapy, especially in treating a lifelong disease such as atherosclerosis. For example, we previously reported that HDAdCMV-gApoAI, which expresses rabbit apolipoprotein A-I (apo A-I) from the cytomegalovirus (CMV) promoter, could retard development of atherosclerosis in rabbit carotid arteries during a 1-month interval.11 However, a 70% decline in apo A-I expression within one month after gene transfer raised concerns about the long-term efficacy of this approach.
Here we tested several strategies aimed either at increasing HDAd-mediated apo A-I expression in EC at all time points or at stabilizing apo A-I expression between day 3 and day 28 after gene transfer (after day 28 HDAd appears to express apo A-I indefinitely in EC).11–13 We first sought to increase translation of transgene mRNA by using species-specific codon-optimized transgene sequences. We next attempted to increase transgene mRNA stability by removing endogenous 3′-untranslated (UTR) sequences. We then tested whether a synthetic mammalian-derived endothelial-selective promoter (4XETE, endothelin-1 [ET-1] enhancer-promoter [ETE] with three additional ETEs added as direct repeats)14 would express apo A-I at higher and more stable levels than the CMV promoter. Finally, we tested the hypothesis that co-expression of interleukin-10 (IL-10) along with apo A-I would increase and stabilize apo A-I expression. This hypothesis was based on our surprising finding that—unlike other transgenes—in vivo HDAd-mediated expression of IL-10 in rabbit EC does not decline over time.15 We found that codon optimization did not improve transgene expression. Complete removal of 3′-UTR sequences substantially increased HDAd-mediated IL-10 expression but eliminated HDAd-mediated apo A-I expression. We also found that both substitution of 4XETE for CMV and co-expression of IL-10 eliminated the early (3- to 28-day) decline of in vivo transgene expression found with HDAdCMV-gApoAI. However, both approaches yielded stable expression by lowering early (3-day) expression of apo A-I rather than raising 28-day expression. It remains a challenge to increase HDAd-mediated apo A-I expression above levels obtained 3 days after infusion of HDAdCMV-gApoAI and to maintain those high expression levels.
Materials and Methods
Construction of 4XETEgApoAI-oPRE expression cassette and HDAd vectors
We previously cloned the rabbit APOAI gene (including 165 bp of 5′-UTR, all apo A-I exons and introns, and 52 bp of 3′-UTR) and ligated it into the plasmid pCIgApoAI.11 The CMV promoter (830 bp; originally from the pCI plasmid; Promega, Madison, WI) was excised from this plasmid by digestion with BglII and NheI. The free ends of the plasmid were blunt-ended with Klenow fragment. The murine endothelin-1 (m-ET1) promoter was removed from plasmid pqET-114 and ligated in place of the CMV promoter to generate pmET1gApoAI.
The APOAI gene sequences described above were then removed from pmET1gApoAI by digestion with SalI and NotI and blunt-ended with Klenow fragment. The plasmid pBshuttle-4XETE-cIL-10-optimal posttranscriptional regulatory element14 was digested with PmeI, to remove the IL10 cDNA. The blunt-ended APOAI gene (from pmET1gApoAI) was ligated in place of the IL10 cDNA to yield pBshuttle-4XETE-gApoAI-oPRE. The expression cassette (containing the 4XETE promoter, rabbit APOAI gene, and the optimal posttranscriptional regulatory element [oPRE]) was transferred to the HDAd backbone plasmid pC4HSU (Microbix Biosystems, Toronto, Canada) by homologous recombination in BJ5183 cells, as described previously.14 HDAd virions were generated by transfection of the recombinant plasmid into 293Cre4 cells and amplification with H14 helper virus (Microbix Biosystems).14,16 This new vector was named HDAd4XETE-gApoAI-oPRE (Fig. 1a). Two other HDAd vectors used in this study (HDAdCMV-gApoAI, and HDAdCMV-Null), were reported previously.11,17 For all HDAd used in this study, concentrated viral stocks ranged from 1 to 4 × 1012 viral particles (vp)/mL, helper virus contamination was <1% (0.1–0.8%), and E1A-containing genomes were below one in 1 × 106 viral genomes (essentially undetectable).
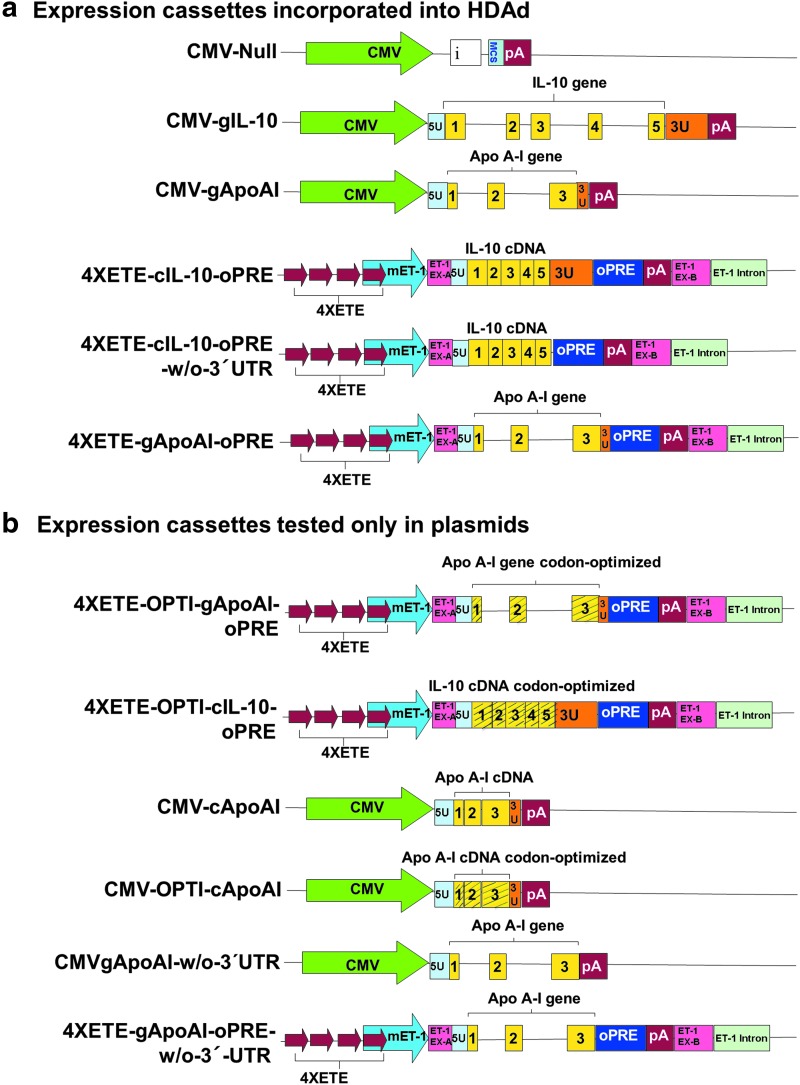
Figure 1.
Expression cassettes (a) incorporated into helper-dependent adenoviral (HDAd) vectors or (b) tested only in plasmids (all of these cassettes were tested after cloning into pBShuttle14). Cassettes incorporated into HDAd were tested first in pBShuttle in vitro and then in HDAd in vitro and in vivo. Drawing is not to scale. Striped yellow blocks (1, 2, 3, 4, and 5) are codon-optimized rabbit interleukin (IL)-10 or apo A-I exons; yellow blocks (1, 2, 3, 4, and 5) are rabbit IL-10 or apo A-I exons; red arrows indicate 4XETE (endothelin-1 [ET-1] enhancer-promoter [ETE] present in ETE with three additional ETEs added as direct repeats). 3U, 3′-untranslated region of either rabbit IL10 or APOAI gene; 5U, 5′-untranslated region of either rabbit IL10 or APOAI gene; CMV, cytomegalovirus promoter from pCI plasmid; ET-1 EX-A and ET-1 EX-B, 5′ and 3′ segments of mouse ET-1 exon 1 (untranslated); ET-1 intron, 5′ segment of mouse ET-1 intron 1; i, β-globin/immunoglobin-G chimeric intron from pCI; IL, interleukin; MCS, multiple cloning site; mET-1, murine endothelin-1 enhancer-promoter; oPRE, optimal woodchuck hepatitis virus post-transcriptional regulatory element; pA, SV40 poly A signal.
Construction of expression cassettes with codon-optimized APOAI and IL10 sequences
Oryctolagus cuniculus-specific codon optimization of the APOAI and IL10 exon sequences was done using proprietary algorithms (GenScript, Piscataway, NJ). A Kozak sequence “CCACC” was introduced upstream of the start codon for all codon-optimized and non-codon-optimized sequences. Codon-optimized sequences of APOAI and IL10 were inserted in place of the corresponding native genomic sequences in pBShuttle-4XETE-gApoAI-oPRE and pBShuttle-4XETE-cIL-10-oPRE, respectively (Fig. 1a). This was accomplished in several steps: (1) Fragments of DNA extending from the SgrAI site in the 4XETE promoter to the BSiWI site in the oPRE (including part of the 4XETE promoter, Kozak sequence, codon-optimized APOAI or IL10, SV40 poly A signal, and part of the oPRE) were synthesized by GenScript and assembled within pUC57 plasmids; (2) fragments containing the codon-optimized APOAI and IL10 sequences were then removed from pUC57 by digestion with SgrAI and BsiWI; and (3) the fragments were ligated to SgrAI/BsiWI-digested pBShuttle-4XETE-gApoAI-oPRE or pBShuttle-4XETE-cIL-10-oPRE, respectively. The ligation products were termed pBShuttle-4XETE-OPTI-gApoAI-oPRE and pBShuttle-4XETE-OPTI-cIL-10-oPRE (Fig. 1b). Presence of codon-optimized sequences was confirmed by DNA sequencing.
Codon optimization of rabbit cDNA encoding apo A-I was also done using proprietary algorithms (GeneArt, Grand Island, NY). The codon-optimized sequence was used to replace the native APOAI cDNA sequence in pBShuttle-CMV-cApoAI (Fig. 1b). This was accomplished in several steps: (1) Fragments of DNA extending from the SnaBI site in the CMV promoter to the HpaI site in the SV40 poly A signal (including part of the CMV promoter, Kozak sequence, codon-optimized APOAI cDNA, and part of SV40 poly A signal) were synthesized and assembled into a pUC57 plasmid; (2) a fragment containing the codon-optimized APOAI cDNA was removed from pUC57 by digestion with SnaBI and HpaI; and (3) the fragment was ligated to SnaBI/HpaI -digested pBShuttle-CMV-cApoAI, replacing the non-codon optimized APOAI cDNA with a codon-optimized version. The ligation product was termed pBShuttle-CMV-OPTI-cApoAI. Presence of codon-optimized sequences was confirmed by DNA sequencing. All codon-optimized sequences are in Supplementary Fig. S1 (Supplementary Data are available online at www.liebertpub.com/hum).
Deletion of IL10 and APOAI 3′-UTR sequences
We deleted 446 bp of native rabbit IL10 3′-UTR sequence from pBshuttle-4XETE-cIL-10-oPRE by using site-directed mutagenesis to insert MluI restriction sites upstream and downstream of the 3′-UTR (QuikChange Lightning Site-Directed Mutagenesis Kit; Agilent Technologies, Santa Clara, CA). The plasmid was then digested with MluI, and re-ligated to yield a new plasmid: pBshuttle-4XETE-cIL-10-oPRE-w/o-3′UTR (Fig. 1a). The expression cassette from this plasmid was inserted into the HDAd backbone plasmid pC4HSU and HDAd4XETE-cIL-10-oPRE-w/o-3′UTR virions were generated as described above. Essentially the same approach was used to remove APOAI 3′-UTR sequences (51 bp) from both pBshuttle-4XETE-gApoAI-oPRE and pBshuttle-CMV-gApoAI (Fig. 1a). The plasmid products were named pBshuttle-4XETE-gApoAI-oPRE-w/o-3′-UTR and pBshuttle-CMV-gApoAI-w/o-3′UTR, respectively (Fig. 1b).
IL-10 and apo A-I expression in vitro
Bovine aortic endothelial cells (BAEC) were purchased (Cell Applications Inc., San Diego, CA). Cells were grown in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Passages 6–8 were used for experiments. We used BAEC because we can no longer obtain rabbit EC. For experiments that required transfection into rabbit cells (i.e., species-specific codon optimization experiments), we used rabbit skin fibroblasts. Fibroblasts were grown as explant cultures from New Zealand White rabbit skin removed postmortem. Cells were frozen in liquid nitrogen at passage 5, then thawed, expanded in DMEM with 10% FBS, and used for experiments at passage 8. Cells were transduced by addition of HDAd diluted in DMEM with 10% FBS to concentrations of 1–2 × 1010 vp/mL (multiplicity of infection approximately 2 × 104). Alternatively, cells were transfected with plasmid DNA using the jetPRIME transfection reagent according to the manufacturer's protocol (Polyplus, New York, NY). Five hours later, the cells were washed twice with DMEM and 600 μL of DMEM was added to the wells. Nineteen hours later, conditioned medium was collected for protein analysis and cells were harvested for RNA and DNA quantitation.
Cellular RNA and DNA were purified using the AllPrep DNA/RNA mini kit (Qiagen, Valencia, CA). RNA was digested with DNase I to remove contaminating genomic DNA. RNA (100 ng) was reverse-transcribed and amplified with a one-step quantitative real-time PCR protocol using qRT-PCR reagents from Thermo Scientific (Grand Island, NY). HDAd vector copies were measured by real-time PCR using ABsolute™ Probe-Based Detection qPCR Master Mix (Thermo Scientific). Apo A-I and IL-10 mRNA levels were normalized to the number of vector copies in the same cells. PCR primers and protocols were as reported for apo A-I11 and IL-10.14
Apo A-I and IL-10 proteins were detected by western blotting of conditioned medium, essentially as decribed.11,14 The only exception was that the secondary antibody used in this study was different (SC-2033; donkey anti-goat immunoglobin-G conjugated to horseradish peroxidase (SantaCruz, Dallas, TX). Protein levels were normalized to plasmid or viral DNA in cell extracts, as described.14
To test whether vector-produced rabbit IL-10 protein altered expression of apo A-I from HDAd we generated IL-10-containing and control conditioned media by transducing BAEC in 6-well plates with either HDAd4XETE-cIL-10-oPRE or HDAdCMV-Null (1 × 1010 pt/mL) for 6 hr. The medium was aspirated, the cells were rinsed three times with DMEM, and fresh DMEM medium was added. After 24 h, the conditioned medium was collected and frozen at –80°C. We transduced other BAEC for 6 h with either HDAdCMV-gApoAI or HDAd4XETE-gApoAI-oPRE (1 × 1010 pt/mL). The medium was aspirated and the cells were rinsed three times with DMEM. The cells were then fed with medium conditioned by BAEC transduced with either HDAd4XETE-cIL-10-oPRE (IL-10-containing medium) or HDAdCMV-Null (control medium). The newly fed BAEC were harvested 24 h later and RNA was extracted for measurement of apo A-I and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA.
In vivo gene transfer to rabbit carotid arteries
All animal protocols were approved by the Office of Animal Welfare of the University of Washington. Male New Zealand White rabbits (3–4 kg, Western Oregon Rabbit Co.) were fed normal chow for one week, and surgically isolated common carotid arteries were infused with HDAd vectors, as reported.18–20 Carotid arteries were harvested 3 or 28 days after gene transfer and cut into five equal segments designated A, B, C, D, and E (cranial to caudal). Segments A, C, and E were snap-frozen in liquid nitrogen for later extraction of RNA with the RNeasy mini kit (Qiagen). Segments B and D (used for protein and DNA analyses) were placed into 1 mL DMEM.
IL-10 and apo A-I expression in vivo
Artery segments B and D were placed with DMEM in individual wells in a 48-well plate, then incubated at 37°C with 5% CO2 for 3 h. The segments were rinsed three times with brief agitation in 500 μL DMEM, then incubated an additional 3 h in 1 mL DMEM. Segments were then rinsed three times in 500 μL DMEM with brief agitation and placed in a single well of a 96-well plate with 100 μL DMEM and again incubated at 37°C with 5% CO2. After 20 h, the media and vessel segments were placed into separate tubes and frozen at −80°C for protein and DNA analyses, respectively. To measure HDAd vector copy number, DNA was isolated from each of the segments, using the DNeasy Blood and Tissue kit (Qiagen). Total RNA extracted from segments A, C, and E of the carotid arteries was quantified by Nanodrop spectrophotometer (Thermo Scientific; Wilmington, DE). Apo A-I and IL-10 mRNA were measured by quantitative reverse transcriptase-mediated PCR, as described above.
Selection of a “housekeeping” gene for normalizing transgene expression
As suggested,21–23 we used a panel of candidate “housekeeping” genes to develop a strategy for normalizing transgene (apo A-I and IL-10) expression in artery extracts. We purchased primers for 6 candidate reference genes (PPIA, ACTB, YWHAZ, HPRT1, SDHA, and CANX) from Primerdesign Ltd. (Southampton, United Kingdom; sequences not disclosed to us). We used these primers, along with our own GAPDH primers,11 to measure expression of GAPDH as well as these six candidate reference genes in eight samples from each group in an in vivo experiment comparing expression of apo A-I from HDAdCMV-gApoAI and HDAd4XETE-gApoAI-oPRE. We also measured expression of GAPDH and the six candidate reference genes in 18 samples from each group in an in vivo experiment comparing the effect of coinfusion of HDAd4XETE-cIL-10-oPRE or HDAdCMV-Null on the expression of apo A-I from HDAdCMV-gApoAI. We used the website RefFinder (compares algorithms from BestKeeper, NormFinder, GeNorm, and the comparative delta-Ct method) and the updated GeNorm algorithm (QBase+) to analyze reference gene expression in samples from these two experiments. These algorithms differed as to their selection of the best reference gene(s), suggesting that there was not a single “best” reference gene or combination. Moreover, use of any of these algorithms for normalization of either data set yielded results that were qualitatively identical. Some authors report that the rRNA:mRNA ratio is inconsistent within experimental samples, and discourage normalization to 18S rRNA.22 However, using a subset of the samples from one of the experiments, we found that normalization of apo A-I expression to 18S rRNA yielded results consistent with those obtained by normalizing to the mRNA reference genes.
Normalization of experimental gene expression to expression of one or more reference genes in the same samples should decrease data variance and thereby improve statistical efficiency.23 Surprisingly, we found that use of the GeNorm-selected reference genes increased the variability (and therefore lowered the statistical efficiency) of our gene-expression data compared with normalizing to GAPDH alone. Because none of the reference gene algorithms altered our results qualitatively and at least one of them increased the data variability, we decided to continue the economical and simple practice of normalizing transgene expression to GAPDH expression in the same samples.11,14,15
Statistics
Two groups were compared by t-test if conditions of normality and equal variance were met; otherwise we used the Mann-Whitney rank-sum test. We used two-way ANOVA to test the effects of time and 3′-UTR on IL-10 expression after plasmid transfection. For experiments with four groups we typically used two-way ANOVA with the Holm-Sidak method for post-hoc pairwise multiple comparisons. For some experiments with four groups, our hypotheses were confined to only two of the groups. In these cases two-group comparisons were performed as above. In one experiment with four groups and P ≥ 0.2 by 2-way ANOVA, we performed post-hoc t-tests to more firmly establish a lack of pairwise differences. In an experiment with four groups in which conditions of normality and equal variance were not met, we used linear regression to test for significant interactions between the two experimental factors.
Results
Codon optimization does not improve expression of IL-10 or apo A-I
We tested codon optimization as a strategy for increasing transgene expression at all time points. Optimization of codon sequences within exons of the rabbit APOAI gene (by GenScript) resulted in synonymous substitutions of 108 out of 267 codons (40.5%). Codon optimization of the rabbit APOAI cDNA (by GeneArt) resulted in 93 synonymous codon substitutions (34.8%). Codon optimization of the rabbit IL10 cDNA (by GenScript) resulted in synonymous substitutions of 87 of 178 codons (48.9%). Calculated codon adaptation index24,25 (1.0 = “perfect”; ≥0.8 = “good”) was 0.84 for native APOAI and increased to 0.85 and 0.95 for the APOAI genomic clone and cDNA, respectively. IL-10 codon adaptation index increased from 0.73 to 0.87. Percent GC content (high GC content can yield RNA secondary structure that interferes with translation) was either essentially the same or slightly less in codon-optimized sequences: 63.1% in native APOAI versus 58% for the codon-optimized APOAI genomic clone and 55% for the codon-optimized APOAI cDNA; 55.1% for native IL-10 versus 56.8% for codon-optimized IL10 cDNA.
We tested the efficacy of codon optimization by transfecting plasmids expressing either the native or codon-optimized sequences (Fig. 1) into cultured rabbit fibroblasts. Apo A-I and IL-10 protein were measured by western blot and normalized to the amount of plasmid DNA in extracts of the transfected cells. Surprisingly, codon optimization did not increase expression of either apo A-I or IL-10 protein (Supplementary Figs. S2, S3).
Complete removal of 3′ UTR sequences has opposite effects on expression of IL-10 and apo A-I
We transfected plasmids containing the rabbit IL10 cDNA either with or without the 5′-most 446 bases of the 3′-UTR (Fig. 1a; compare 4XETE-cIL-10-oPRE and 4XETE-cIL-10-oPRE-w/o-3′UTR) into cultured bovine aortic endothelial cells (BAEC) and measured IL-10 mRNA 24, 48, 72, and 96 h later. We anticipated that absence of the 3′ UTR would remove miRNA binding sites that contribute to mRNA silencing or degradation, thereby increasing IL-10 mRNA levels at all time points.26 Removal of the 3′-UTR sequences significantly increased IL-10 mRNA expression at early but not at late time points (P = 0.001; Fig. 2a). Based on this encouraging result, we attempted the same strategy with apo A-I. We removed the residual 51 bases of the endogenous APOAI 3′-UTR from both pBshuttle-4XETE-gApoAI-oPRE and pBshuttle-CMV-gApoAI (Fig. 1a, b; compare 4XETE-gApoAI-oPRE and 4XETE-gApoAI-oPRE-w/o-3′UTR as well as CMV-gApoAI and CMV-gApoAI-w/o-3′UTR). We transfected plasmids with each of these 4 expression cassettes into BAEC. Surprisingly, in both cases removal of the 3′-UTR sequences resulted in near-complete loss of apo A-I mRNA. Apo A-I mRNA decreased by 94% after removal of the 3′-UTR sequences from pBshuttle-4XETE-gApoAI-oPRE and by 91% after removal of the 3′-UTR sequences from pBshuttle-CMV-gApoAI (P = 0.002 for both; data not shown).
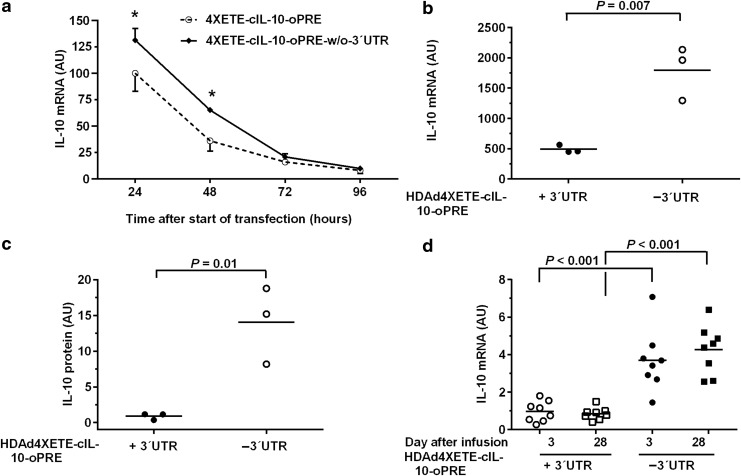
Figure 2.
Increased IL-10 expression after removal of 3′ UTR. (a) Bovine aortic endothelial cells (BAEC) were transfected either with a plasmid (Fig. 1) containing 446 bp of the native IL10 3′ UTR (pBshuttle-4XETE-cIL-10-oPRE) or with a plasmid lacking any 3′ UTR sequences (pBshuttle-4XETE-cIL-10-oPRE-w/o-3′UTR). IL-10 mRNA was measured by qRT-PCR 24, 48, 72, and 96 hours after transfection. IL-10 expression is calculated as a percentage of the mean expression from the 3′ UTR-containing vector at 24 hours (arbitrarily set to 100%). Data are mean and SD; n = 3; *P < 0.001 versus UTR-containing vector; two-way analysis of variance (ANOVA) with post-hoc correction for pairwise comparisons. (b) BAEC were transduced with HDAd-4XETE-cIL-10-oPRE vectors either with the IL10 3′ UTR sequences (+ 3′UTR) or without the 3′ UTR (–3′UTR). IL-10 mRNA was measured 24 hours after transduction by qRT-PCR and normalized to GAPDH mRNA and vector DNA in cell extracts. (c) IL-10 protein was measured in conditioned medium of the same cells as in (b) by western blot, also at 24 hours. The density of bands was normalized to vector DNA in cell extracts. For (b) and (c), data points are individual wells; bars indicate means; P values are from t-tests. (d) Rabbit EC were transduced in vivo by infusion of same HDAd vectors used in (b) and (c). 3 and 28 days later, transduced carotids were removed, RNA extracted, and IL-10 mRNA measured by qRT-PCR with normalization to GAPDH mRNA in the same extracts. Data points are individual arteries; bars are means. (d) P values are from two-way ANOVA with post-hoc correction for pairwise comparisons.
We hypothesized that the plasmid transfection results (Fig. 2a) underestimated the impact of 3′-UTR-removal on IL-10 expression because the plasmid and mRNA are present only transiently. We therefore incorporated the 3′-UTR-deleted IL-10 expression cassette into an HDAd vector (HDAd4XETE-cIL-10-oPRE-w/o-3′UTR; Fig. 1a) and compared it to the parent HDAd 4XETE-cIL-10-oPRE vector14 (Fig. 1a) in vitro in BAEC and in vivo in rabbit carotid arteries. In vitro, IL-10 mRNA was increased by 3.5-fold in cells transduced with the vector lacking the 3′-UTR (P = 0.007; Fig. 2b) and protein expression was increased by ∼18-fold (P = 0.01; Fig. 2c, Supplementary Fig. S4). In vivo, deletion of the 3′-UTR increased IL-10 mRNA 4- to 5-fold at both 3 days and 28 days after gene transfer (P < 0.001 for both; Fig. 2d). We could not reliably detect IL-10 protein in the artery explant culture medium, precluding comparison of protein levels. This experiment also revealed that the remarkable early stability (i.e., between 3 and 28 days after transduction) of in vivo expression that we reported for HDAdCMV-gIL-1015 was present for both HDAd4XETE-cIL-10-oPRE-w/o-3′UTR and the parent HDAd 4XETE-cIL-10-oPRE vector (Fig. 2d).
Co-expression of IL-10 in vivo results in stable apo A-I expression between 3 and 28 days
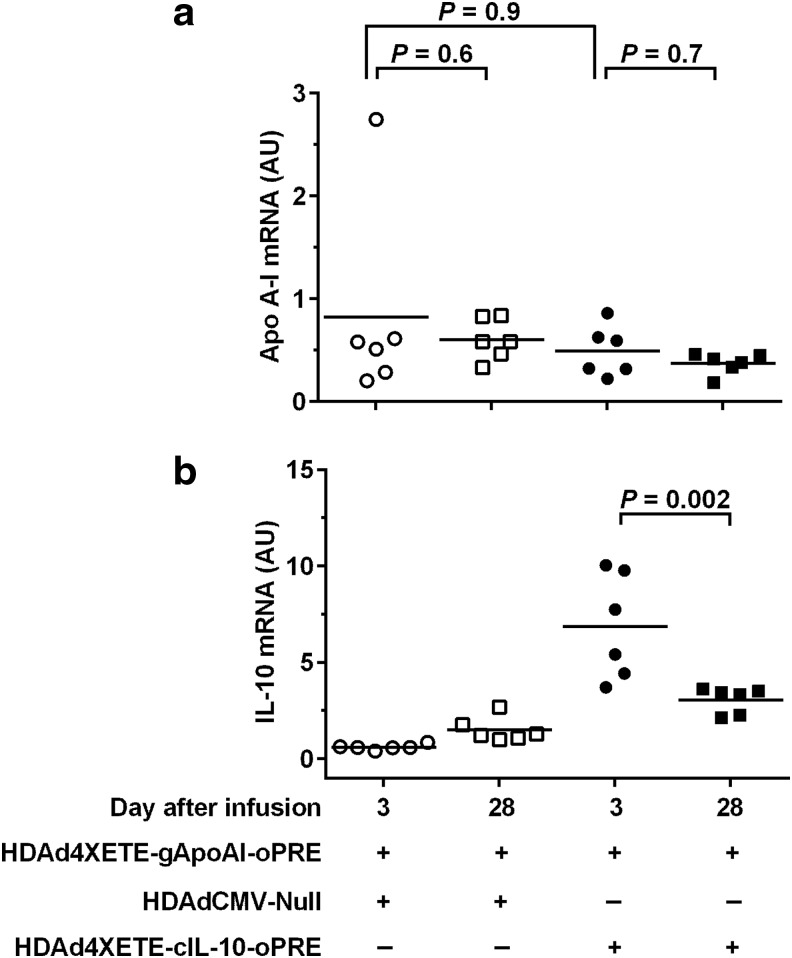
In contrast to the stable-to-increasing in vivo expression of IL-10 from HDAd (whether expressed from the CMV or 4XETE promoter), expression of apo A-I from HDAdCMV-gApoAI (in a previous study) declined ∼70% between 3 and 28 days after infusion into rabbit carotids.11 We hypothesized that IL-10 prevented early loss of IL-10 transgene expression via local anti-inflammatory effects.15,27,28 Accordingly, we tested whether co-expression of IL-10 in arteries transduced with HDAdCMV-gApoAI would stabilize and possibly increase apo A-I expression between 3 and 28 days. An advantage of HDAd is that it has a large cloning capacity (>35 kilobases) that could potentially accommodate expression cassettes for both apo A-I and IL-10. However, in this initial study we used two separate vectors (both of which were already proven in our previous work)11,14 to test this concept. We infused HDAdCMV-gApoAI along with either HDAd4XETE-cIL-10-oPRE or HDAdCMV-Null (as a control). All vectors were infused at a concentration of 2 × 1011 pt/mL (total vector concentration = 4 × 1011 pt/mL: twice the total vector concentration used previously).11,15 Co-infusion of HDAdCMV-gApoAI with HDAdCMV-Null resulted in a large decline in apo A-I transgene expression between 3 and 28 days (79% decrease; P < 0.001; Fig. 3a). In contrast, co-infusion of HDAdCMV-gApoAI with HDAd4XETE-cIL-10-oPRE eliminated the significant decline in apo A-I expression between 3 and 28 days (P = 0.4). Improved stability of apo A-I expression in HDAd4XETE-cIL-10-oPRE co-transduced arteries is more apparent when apo A-I levels are normalized to 100% at day 3 in both groups (Fig. 3b). However, compared to arteries co-infused with HDAdCMV-Null, co-infusion of HDAd4XETE-cIL-10-oPRE was associated with lower expression of apo A-I from HDAdCMV-gApoAI 3 days after gene transfer (58% decrease; P < 0.001; Fig 3a) and co-infusion of HDAd4XETE-cIL-10-oPRE (versus co-infusion of HDAdCMV-Null) did not significantly increase apo A-I at 28 days (P = 0.4).
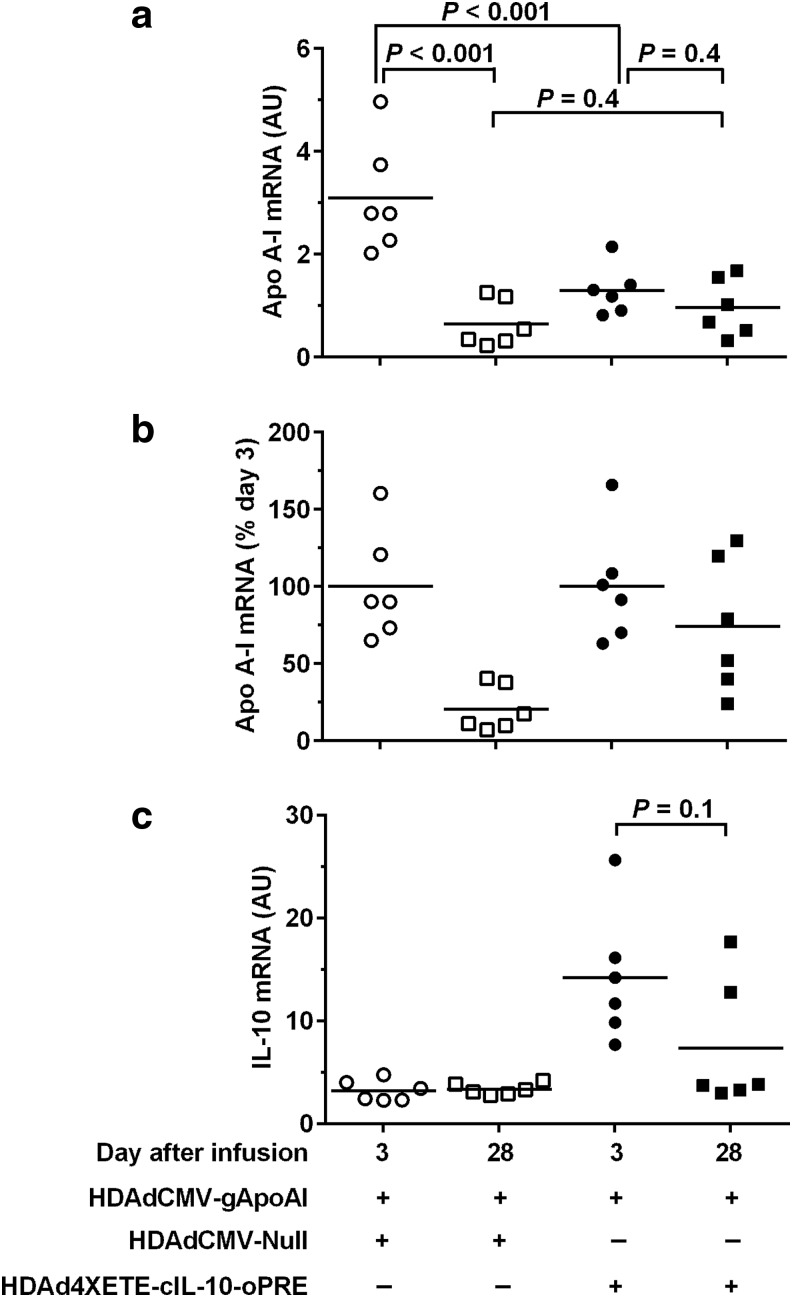
Figure 3.
Co-expression of IL-10 in vivo stabilizes apo A-I expression between 3 and 28 days. Rabbit EC were transduced in vivo by infusion of the indicated vectors. Each vector was infused at 2 × 1011 pt/mL (total 4 × 1011 pt/mL per infusion). Transduced carotids were removed after 3 days and 28 days, and apo A-I and IL-10 RNA were measured by qRT-PCR. (a) apo A-I mRNA (normalized to GAPDH). (b) Data from (a), expressed as percentage of mean day-3 values for each vector. (c) IL-10 mRNA (normalized to GAPDH). Data points are individual arteries; bars are means. (a) P values are from 2-way ANOVA with post-hoc correction for pairwise comparisons. (c) P value is from Mann–Whitney rank-sum test.
To help interpret these results, we measured IL-10 expression in the same arteries. As expected, 3 days after gene transfer, IL-10 mRNA was increased in all arteries co-infused with HDAd4XETE-cIL-10-oPRE (Fig. 3c). By 28 days, IL-10 expression dropped by 48%. Although this drop was not statistically significant (P = 0.1), it differs from the stable or increasing IL-10 expression we observed when an IL-10-expressing vector is infused alone, both in this study (Fig. 2d) and previously.15 Because our finding of increased IL-10 expression between 3 and 28 days in arteries infused with HDAdCMV-gIL-10 (Fig. 1a; Du et al., 201115) provided an important rationale for the IL-10/apo A-I co-infusion studies, we repeated the original (published) experiment comparing in vivo expression from HDAdCMV-gIL-10 at days 3 and 28. We again found that IL-10 expression increased between 3 and 28 days (70% increase; P = 0.06; Supplementary Fig. S5), reproducing this important precedent.
A 4XETE promoter–containing expression cassette stably expresses apo A-I in vivo
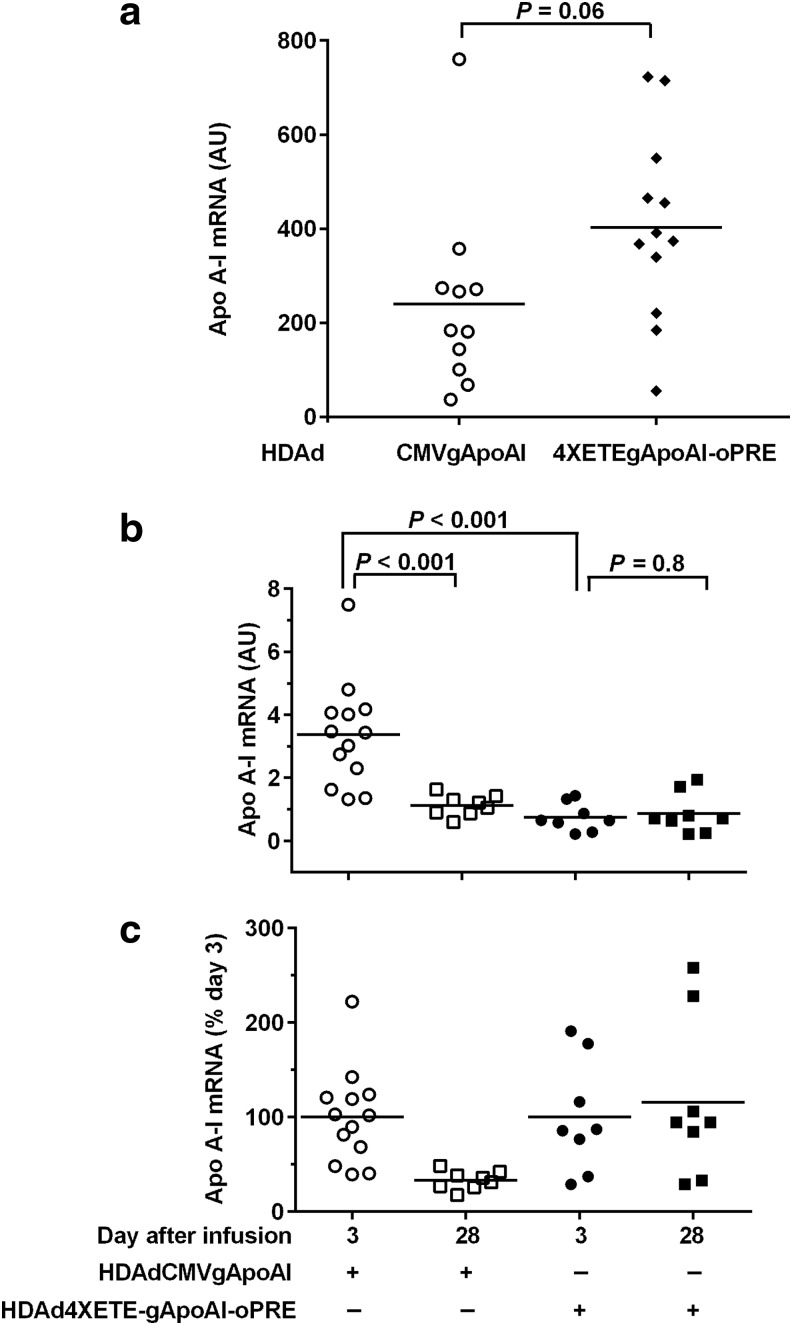
In parallel to the IL-10 co-infusion experiments, we tested whether use of our expression cassette containing the 4XETE promoter14 (which is present in 4XETE-cIL-10-oPRE; Fig. 1a) instead of the CMV promoter would also stabilize in vivo expression of apo A-I between 3 and 28 days. We reported previously that this 4XETE promoter-containing cassette expressed IL-10 at levels at least as high as the CMV promoter both in vitro and in vivo, 3 days after infusion.14 Moreover, because it is a nonviral promoter, 4XETE might be less susceptible to promoter attenuation in vivo.29,30 BAEC transduced with HDAd4XETE-gApoAI-oPRE expressed approximately 70% higher levels of apo A-I mRNA than BAEC transduced with HDAdCMV-gApoAI (P = 0.06; Fig. 4a). In rabbit carotids (in vivo), however, apo A-I expression from HDAd4XETE-gApoAI-oPRE at 3 days after infusion was significantly lower than expression from HDAdCMV-gApoAI (78% less; P < 0.001; Fig. 4b). As expected, between 3 and 28 days, expression from HDAdCMV-gApoAI declined by 70% (P < 0.001). In contrast, expression from HDAd4XETE-gApoAI-oPRE was stable between 3 and 28 days (P = 0.8). Stable expression from HDAd4XETE-gApoAI-oPRE is more apparent when apo A-I levels are normalized to 100% at day 3 in both groups (Fig. 4c). Therefore, both co-infusion of an IL-10 vector (when apo A-I is expressed from the CMV promoter; Fig. 3a) and use of a 4XETE promoter to express apo A-I (Fig. 4b) yield stable apo A-I expression between 3 and 28 days, but both approaches yield lower apo A-I expression 3 days after infusion.
Figure 4.
A 4XETE promoter-containing expression cassette expresses apo A-I at least as well as a CMV promoter-containing expression cassette in vitro and achieves lower, but stable apo A-I expression in vivo. (a) BAEC were transduced with the indicated vectors. Apo A-I mRNA was measured by qRT-PCR 24 hours after transduction and normalized to GAPDH and vector DNA in cell extracts. Data points are individual wells; bars are means; P value is from t-test. (b) Rabbit EC were transduced in vivo by infusion of the same vectors, both at 2 × 1011 pt/mL. Transduced carotid arteries were removed after 3 and 28 days and apo A-I mRNA was measured and normalized to GAPDH mRNA in the same extracts. (c) Data from (b), expressed as percent of mean day 3 values for each vector. (b) and (c) Data points are individual arteries; bars are means. (b) P values are from two-way ANOVA with post-hoc correction for pairwise comparisons.
Co-expression of IL-10 does not decrease apo A-I expression from a 4XETE promoter–containing expression cassette
We hypothesized that lower apo A-I expression 3 days after co-infusion of HDAdCMV-gApoAI with HDAd4XETE-cIL-10-oPRE (Fig. 3a) was due to local anti-inflammatory effects of IL-10 that suppressed early (3-day) expression from the CMV promoter. This hypothesis was based on four premises: (1) the likelihood that a local inflammatory response occurs after open surgical manipulation of a carotid artery; (2) others' data showing that expression from the CMV promoter is upregulated by inflammatory signals;31,32 (3) our own data showing that expression from a CMV promoter in HDAd is upregulated (more than 50-fold) in vivo by co-infusion of a first-generation Ad vector and ex vivo by treatment of transduced arteries with phorbol myristate acetate (six-fold; both first-generation Ad and phorbol myristate acetate are potent inflammatory stimuli);12 and (4) the well-established anti-inflammatory activities of IL-10, which include inhibition of local inflammation after adenoviral vector infusion.27,28 Accordingly, we hypothesized that local IL-10 expression would not suppress expression of apo A-I from the 4XETE promoter, because this promoter is less likely to be upregulated by local inflammatory signals than the CMV promoter. To test this hypothesis, we compared apo A-I expression from 4XETE-gApoAI-oPRE-infused arteries that were co-infused with either HDAdCMV-Null or HDAd4XETE-cIL-10 (total vector concentration 4 × 1011 pt/mL). Compared to arteries co-infused with HDAdCMV-Null, co-infusion of HDAd4XETE-cIL-10-oPRE with HDAd4XETE-gApoAI-oPRE did not decrease apo A-I at 3 days after gene transfer (P = 0.9; Fig. 5a). Moreover, expression of apo A-I from HDAd4XETEgApoAI was stable whether co-infused with HDAdCMV-Null or HDAd4XETE-cIL-10-oPRE (P = 0.4 for an effect of time on apo A-I expression by two-way ANOVA; P ≥ 0.6 for t-tests comparing apo A-I expression at 3 and 28 days; Fig. 5a). To help interpret these results, we measured IL-10 expression from the transduced arteries. Similar to results obtained in the previous co-infusion experiment (Fig. 3c), IL-10 expression in arteries infused with HDAd4XETE-cIL-10-oPRE was elevated at 3 days, but fell significantly by 28 days (55% decrease; P = 0.002; Fig. 5b).
Figure 5.
Co-expression of IL-10 does not decrease in vivo apo A-I expression from a 4X-ETE promoter-containing expression cassette. Rabbit EC were transduced in vivo by co-infusion of the indicated vectors, at 2 × 1011 pt/mL each (total 4 × 1011 pt/mL per infusion). Transduced carotid arteries were removed after 3 and 28 days. (a) apo A-I mRNA and (b) IL-10 mRNA, both normalized to GAPDH mRNA in the same extracts. Data points are individual arteries; bars are means. (a) Two-way ANOVA showed no effect of either IL-10 or time on apo A-I mRNA levels (P ≥ 0.2); P values are from t-tests. (b) P value is from Mann–Whitney rank-sum test.
Increased IL-10 expression lowers apo A-I expression from HDAdCMV-ApoAI in vivo
This second co-infusion experiment supported our hypothesis that the apparent stabilization of apo A-I expression either by co-expression of IL-10 (Fig. 4a) or by substitution of the 4XETE promoter for the CMV promoter (Fig. 5b) was due primarily to decreased apo A-I expression at day 3 rather than increased apo A-I expression at day 28. However, in the first IL-10 co-expression experiment (Fig. 3) IL-10 levels decreased between day 3 and day 28 (Fig. 3c). This drop in IL-10 expression contrasted with our findings that HDAd-mediated IL-10 expression either remained stable or increased from 3 to 28 days when an IL-10-expressing vector is infused alone (Fig. 2d, Supplementary Fig. S5, and Du et al.15). This discrepancy left open the possibility that achievement of stable or elevated IL-10 expression levels at 28 days would boost expression from a co-infused apo A-I–expressing vector.
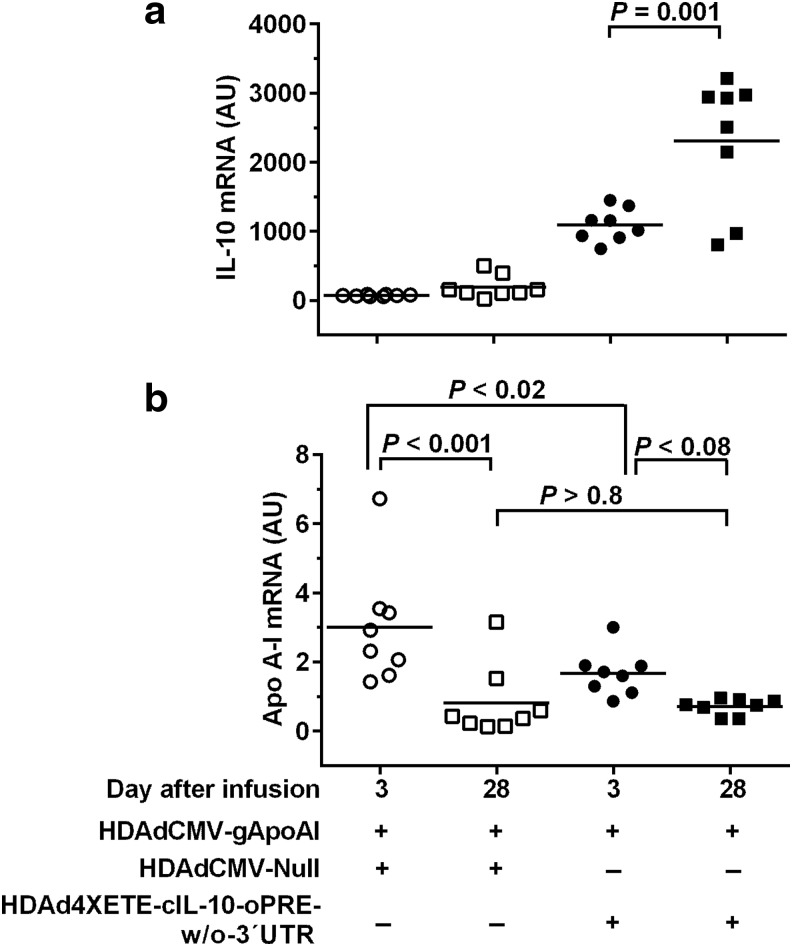
We considered the possibility that the drop in IL-10 expression between days 3 and 28 in the co-infusion experiment (versus stability of IL-10 expression when an IL-10-expressing vector is infused alone) was due to toxicity associated with use of a two-fold higher total vector dose in the co-infusion experiments. To address this, while attempting to preserve expression of both apo A-I and IL-10, we lowered the HDAdCMV-gApoAI dose by 25% (to 1.5 × 1011 pt/mL) and substituted the higher-expressing HDAd4XETE-cIL-10-oPRE-w/o-3′UTR vector (at 0.5 × 1011 pt/mL) for HDAd4XETE-cIL-10-oPRE. This strategy lowered the total vector dose to 2 × 1011 pt/mL, a concentration associated with stable or increased expression of IL-10 at 28 days (Fig. 2d, Supplementary Fig. S5, and Du et al., 201115). This new co-infusion strategy prevented loss of IL-10 expression: IL-10 mRNA levels increased between 3 and 28 days (110%; P = 0.001; Fig. 6a). However, compared to co-infusion of HDAdCMV-Null (also at 0.5 × 1011 pt/mL), co-infusion of HDAd4XETE-cIL-10-oPRE-w/o-3′UTR tended not to stabilize apo A-I expression from HDAdCMV-gApoAI between 3 and 28 days (58% drop; P < 0.08; Fig. 6b). Moreover, apo A-I levels were not increased in 28-day arteries with increased IL-10 expression (P > 0.8 compared with 28-day arteries co-infused with HDAdCMV-Null). Therefore, elevated IL-10 expression at 28 days did not increase HDAdCMV-gApoAI expression between 3 and 28 days. In addition, elevated IL-10 expression at day 3 decreased apo A-I expression from HDAdCMV-gApoAI (44% lower apo A-I expression in HDAd4XETE-cIL-10-oPRE-w/o-3′UTR-co-infused arteries versus HDAdCMV-Null arteries; P < 0.02; Fig. 6b).
Figure 6.
Increased IL-10 expression at 28 days does not increase apo A-I expression from HDAdCMVgApoAI in vivo. Rabbit EC were transduced in vivo by co-infusion of the indicated vectors. HDAdCMVgApoAI was infused at 1.5 × 1011 pt/mL and the other vectors at 0.5 × 1011 pt/mL (total 2 × 1011 pt/mL per infusion). Transduced carotid arteries were removed after 3 and 28 days. (a) IL-10 mRNA and (b) apo A-I mRNA, both normalized to GAPDH mRNA in the same extracts. Data points are individual arteries; bars are means. (a) P value is from Mann–Whitney rank-sum test. (b) P values are from two-way ANOVA with post-hoc correction for pairwise comparisons.
IL-10 does not directly suppress expression from HDAdCMV-gApoAI in vitro
The in vivo data showed that co-transduction with an IL-10-expressing vector does not increase apo A-I expression from HDAdCMV-gApoAI at 28 days; rather, it suppresses apo A-I expression from HDAdCMV-gApoAI at 3 days (Figs. 3a, 6b). This suppression could result from cell-autonomous effects of IL-10 on HDAdCMV-gApoAI-transduced endothelial cells. Alternatively, IL-10 might suppress HDAdCMV-gApoAI expression indirectly, via actions on other cell types, including immune cells. To test the hypothesis that IL-10 protein acts directly on transduced endothelial cells to suppress expression from HDAdCMV-gApoAI, we transduced BAEC with HDAdCMV-gApoAI then treated them with conditioned medium that either contained or lacked rabbit IL-10. Presence/absence of IL-10 protein in conditioned media was confirmed by western blot (data not shown). We performed the same experiment with BAEC transduced with HDAd4XETE-gApoAI-oPRE. IL-10-containing medium had no significant effect on expression from HDAdCMV-gApoAI, and modestly increased expression from HDAd4XETE-gApoAI-oPRE (Supplementary Fig. S6).
Stabilization of Apo A-I gene expression is independent of effects on HDAd genome loss
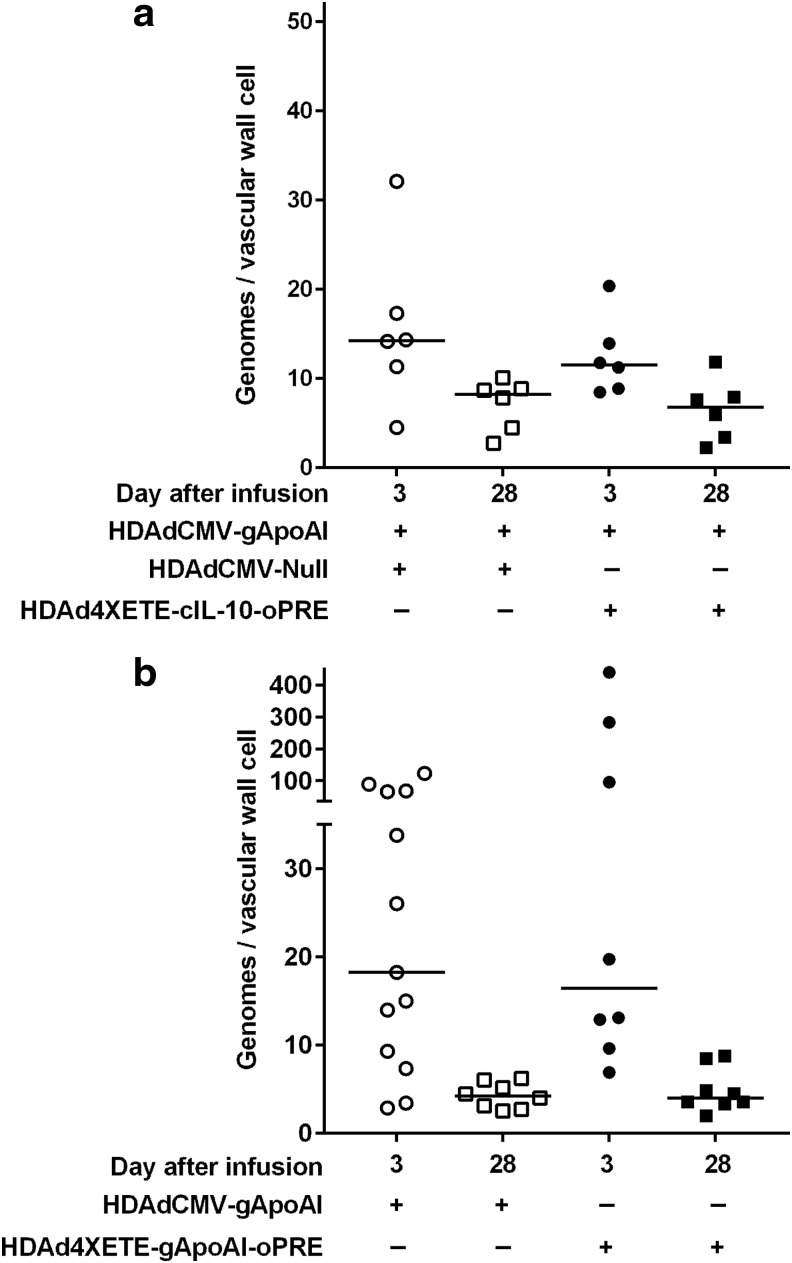
Our initial in vivo experiments with HDAdCMV-gIL-10 suggested that stability of IL-10 mRNA was associated with—and was potentially caused by—a local effect of IL-10 that increased the persistence of HDAd genomes.15 To test the hypothesis that in vivo stability of apo A-I expression in the present studies—either in arteries co-infused with IL-10 (Fig. 3) or in arteries infused with HDAd4XETE-gApoAI-oPRE (Fig. 4)—was associated with improved persistence of HDAd genomes, we measured HDAd genomes in segments from the same 3- and 28-day arteries in which we measured apo A-I mRNA. Neither co-expression of IL-10 nor substitution of the 4XETE expression cassette for the CMV promoter altered the extent of HDAd genome loss between 3 and 28 days. Co-infusion of HDAd4XETE-cIL-10 was associated with a 41% drop in median HDAd genomes at 28 days versus 42% with HDAdCMV-Null (P = 1.0 for an interaction between time and presence of IL-10; Fig. 7a). Loss of genomes between 3 and 28 days was essentially identical for HDAdCMV-gApoAI and HDAd4XETE-gApoAI-oPRE genomes (77% and 76% median loss, respectively; P = 0.5 for an interaction between time and expression cassette; Fig. 7b).
Figure 7.
Neither IL-10 co-transduction nor use of the 4XETE expression cassette stabilizes HDAd genomes. Rabbit EC were transduced in vivo by infusion of the indicated vectors. (a) Each of the vectors was infused at 2 × 1011 pt/mL (total 4 × 1011 pt/mL per infusion). (b) Each vector was infused at 2 × 1011 pt/mL. Transduced carotid arteries were removed after 3 days and 28 days. HDAd genomes were measured by qPCR and normalized to the number of cells represented by total DNA in each extract. Data points are individual arteries; bars are medians. Analysis with linear regression yielded (a) P = 1.0 for interaction between time and presence of IL-10; and (b) P = 0.5 for interaction between time and expression cassette.
Discussion
We tested several strategies aimed either at increasing HDAd-mediated apo A-I expression at all time points or at preventing early loss of apo A-I expression after in vivo gene transfer to EC. We also tested whether two of these strategies could increase expression of IL-10 from HDAd. Our major findings were: (1) Codon optimization did not increase synthesis of either apo A-I or IL-10 protein. (2) Complete removal of 3′-UTR sequences significantly increased IL-10 expression but essentially eliminated apo A-I expression. (3) Co-infusion of IL-10-expressing vectors stabilized in vivo expression from HDAdCMV-gApoAI by lowering apo A-I expression 3 days after gene transfer. (4) Substitution of the 4XETE promoter for the CMV promoter also stabilized in vivo expression from HDAdCMV-gApoAI by lowering apo A-I expression 3 days after gene transfer. (5) Locally expressed IL-10 can significantly downregulate CMV promoter-driven expression—but not 4XETE promoter-driven expression—from HDAd. (6) HDAd-mediated transgene expression in EC is inherently stable in vivo and early (3- to 28-day) declines measured in transgene expression from the CMV promoter are due to upregulation of CMV promoter activity at 3 days. (7) Significant early (3- to 28-day) loss of HDAd genomes does not result in loss of HDAd-mediated transgene expression.
We began by testing whether codon optimization could increase transgene expression. This approach is based on a belief that native DNA sequences do not direct the highest possible levels of protein expression, and that replacing native codons with more optimal codons can facilitate higher, potentially nonphysiologic levels of expression. Approaches for achieving codon optimization are complex, species-specific, and beyond the scope of this article.24,33 Others have reported impressive success with codon optimization,34,35 but there are also failures.36,37 Here we took the common approach of relying on confidential algorithms of commercial DNA synthesis companies, designed in part with a goal of improving the codon adaptation index.25,33 We were uniformly unsuccessful, illustrating the imperfect state of this approach.38 Nevertheless, we acknowledge that other codon-optimization algorithms might be effective in increasing expression of rabbit apo A-I and IL-10.
Our attempts to increase transgene expression by elimination of 3′-UTR sequences yielded mixed results. 3′-UTR sequences are well-known mediators of mRNA degradation that function as targets for miRNA-mediated RNA catabolism.26 Accordingly, many builders of gene therapy vectors routinely delete large segments of endogenous 3′-UTR both for this reason and to accommodate vector-specific limits on genome size. However, 3′-UTR sequences that mediate mRNA stability also exist and incorporation of these sequences can increase transgene mRNA stability and transgene protein expression.26,39,40 A recent study suggests that stabilizing elements are common in human 3′-UTRs;41 our results indicate that the proximal 3′-UTR of rabbit apo A-I contains a strong stabilizing element. Future studies will determine whether insertion of this element into the 3′-UTR of other genes can stabilize mRNA and increase protein expression.
Our efforts to increase vector-driven apo A-I expression by co-expression of IL-10 or by substitution of the 4XETE promoter for the CMV promoter were based on a working model that was derived from our previous experimental data.11,14,15 These efforts also anticipated that co-expression of IL-10 with apo A-I might provide additional atheroprotection, either by direct anti-inflammatory effects of IL-10 or by increased apo A-I-mediated cholesterol efflux.42 Our working model (Supplementary Fig. S7a) postulated that early loss of apo A-I expression from HDAdCMV-gApoAI11 was caused by a combination of downregulated expression from the CMV promoter (either by epigenetic modification29,43 or by cytokine-mediated effects32) and loss of HDAd genomes due to immune-mediated clearance of transduced EC. Because we did not find an early decline in transgene expression after infusion of HDAdCMV-gIL-10 in the same model, and because stable IL-10 expression was associated with modestly increased stability of HDAd genomes,15 we hypothesized that IL-10 acted via local immunomodulatory effects27,28 that either prevented modification/downregulation of the CMV promoter or blocked immune-mediated clearance of transduced cells (Supplementary Fig. S7b). Accordingly, we hypothesized that co-expression of IL-10 would prevent loss of apo A-I expression from HDAdCMV-gApoAI (Supplementary Fig. S7c). Moreover, because the 4XETE promoter is not viral-based, because it expressed IL-10 in vivo at levels at least as high as those obtained with the CMV promoter,14 and because we deleted 5′ sequences that mediate regulation by some inflammatory stimuli from the mouse endothelin promoter fragment included in 4XETE,44–46 we hypothesized that substituting 4XETE for CMV would reduce early loss of transgene expression that occurs either via epigenetic or cytokine-mediated pathways (Supplementary Fig. S7d).
In contrast to our expectations (Supplementary Fig. S7c), the predominant effect of IL-10 co-expression (Fig. 3a, b) was to decrease early (3-day) apo A-I expression from the CMV promoter while leaving 28-day expression levels unaltered (Supplementary Fig. S8a). When IL-10 expression was increased at 28 days by use of a more potent vector (Fig. 6), there was a trend towards reduced expression from HDAdCMV-gApoAI, compared with 3-day expression from the same vector (Supplementary Fig. S8b). The absence of reduced apo A-I expression in this experiment in 28-day arteries cotransduced with the 4XETE-cIL-10-oPRE-w/o-3′UTR vector compared to 28-day arteries cotransduced with HDAdCMV-Null (Fig. 6) could be due either to achievement of maximal IL-10 effects in 28-day IL-10–cotransduced arteries (i.e., IL-10 cannot suppress expression below a certain baseline level) or to promoter competition that concurrently suppressed expression in HDAdCMV-Null–cotransduced EC. Importantly, the suppressive effects of IL-10 on expression from HDAdCMV-gApoAI cannot be attributed to promoter competition between the IL-10 and apo A-I vectors, because promoter competition would be far more likely to occur with the control HDAdCMV-Null vector than with the experimental 4XETE-IL-10 expression cassettes.
The unanticipated suppression of HDAdCMV-gApoAI expression by co-expression of IL-10 in vivo, the lack of suppression of apo A-I expression when IL-10 was co-expressed in vivo with HDAd4XETE-gApoAI-oPRE, and the failure of IL-10-containing CM to suppress apo A-I expression from HDAdCMV-gApoAI-transduced EC in vitro led us to develop a new working model (Supplementary Fig. S8c). We now hypothesize that vector-produced IL-10 acts in vivo on vascular cells to block acute inflammatory effects associated either with the gene transfer surgery itself or with the acute infusion of HDAd particles. These effects—likely cytokine mediated—acutely increase expression from the CMV promoter but not the 4XETE promoter (Supplementary Fig. S8d). Therefore, vector-produced IL-10 prevents cytokine-mediated upregulation of the CMV promoter. This model can explain the results of all of our co-infusion experiments (Figs. 3, 5, 6). This model of cytokine-mediated acute upregulation of the CMV promoter and blockade of this effect by IL-10 may also have important implications for other studies. First, it provides a novel explanation for early declines in transgene expression from the CMV promoter, declines that are often attributed to promoter shutdown.29,43 Second, it suggests that vectors expressing IL-10 may downregulate their own expression if they contain an inflammation-responsive promoter, and that higher levels of IL-10 expression could exacerbate this problem. Co-expression of apo A-I and IL-10—especially when these atheroprotective genes are placed in the same vector—might yet be an effective means of delivering gene therapy for atherosclerosis. However, we no longer anticipate that co-expression of IL-10 would increase or stabilize apo A-I expression.
As with the co-infusion experiments, some of the results we obtained on substituting 4XETE for the CMV promoter were unanticipated. For example, our in vitro results showed that HDAd4XETE-gApoAI-oPRE expressed at least as much apo A-I mRNA as HDAdCMV-gApoAI. Based on this result and on analogous experiments with the IL-10-expressing vectors HDAd4XETE-cIL-10-oPRE and HDAdCMV-cIL-10 (in which HDAd4XETE-cIL-10-oPRE expressed at least as much IL-10 mRNA as HDAdCMV-cIL-10 3 days after in vivo gene transfer), we expected HDAd4XETE-gApoAI-oPRE to express at least as much apo A-I mRNA as HDAdCMV-gApoAI in vivo. However, HDAd4XETE-gApoAI-oPRE expressed far less apo A-I mRNA than HDAdCMV-gApoAI in vivo. On the other hand, our expectation that substitution of the 4XETE promoter for the CMV promoter would stabilize apo A-I expression in vivo was borne out (although stability was at a lower level than expected).
These results with HDAd4XETE-gApoAI-oPRE also led us to modify our working model. We now hypothesize that HDAd containing the 4XETE promoter stably express transgenes in EC in vivo because—unlike the CMV promoter—the 4XETE promoter is not acutely upregulated by local inflammatory events that occur early after gene transfer. This model also explains our observation that a 4XETE-containing cassette—but not a CMV-containing cassette—stably expresses apo A-I in vivo (Supplementary Fig. S8e, f). Our model can also explain why CMV-driven expression of IL-10 increases in vivo between 3 and 28 days (Du et al., 2011,15 and Supplementary Fig. S5); whereas 4XETE-driven expression of IL-10 is stable between 3 and 28 days (Fig. 2d). The model attributes this difference to IL-10 blockade of factors that are present 3 days after gene transfer and that typically upregulate CMV promoter activity but do not upregulate 4XETE promoter activity. Our model fits all results obtained when 4XETE- and CMV-containing apo A-I or IL-10 vectors are infused alone. The model also explains why HDAd4XETE-gApoAI-oPRE expresses at least as much apo A-I mRNA as HDAdCMV-gApoAI in vitro: there is no host inflammatory response in vitro to upregulate expression from the CMV promoter in HDAdCMV-gApoAI. Finally, stable transgene expression from the HDAd4XETE-gApoAI-oPRE between days 3 and 28 argues that significant EC proliferation and consequent dilution/loss of transgene expression does not occur during this interval.
Measurements of vector DNA made in parallel with measurements of transgene expression tested another aspect of our initial model: that loss of transgene expression was due, at least in part, to loss of vector genomes (Supplementary Fig. S7a). Surprisingly, we found that in two settings in which apo A-I transgene expression is completely stable between 3 and 28 days (co-expression of IL-10 and use of the 4XETE promoter) HDAd genomes were lost at the same rate as in parallel experiments in which transgene expression declined by 70–80%. Moreover, when we plot transgene expression versus vector genomes measured in individual arteries harvested at the same time point and containing the same vectors, these 2 measurements are usually only weakly correlated and sometimes are not correlated at all (data not shown). Our revised model fits these new data (compare Supplementary Figs. S7a and S8e; Fig. S8e does not include a role for genome loss). We now hypothesize that a significant percentage of vector genomes in transduced carotid arteries are not transcriptionally active; therefore, their loss does not affect transcription. An HDAd genome could be transcriptionally inactive either because it is inside a virion that is extracellular or because it did not enter the nucleus and become chromatinized.47,48 Our data indicate that loss of HDAd genomes from carotid arteries is an epiphenomenon caused by passage of time, that this genome loss does not impact expression from either the CMV or 4XETE promoters, and that prevention of genome loss should not always be a primary target of efforts to stabilize transgene expression.
We did not achieve our initial goals of increasing apo A-I expression from HDAd and sustaining apo A-I expression at a high level. Nevertheless, there are positive aspects to this study. Most importantly, we discovered that in vivo transduction of EC with HDAd can reproducibly confer stable transgene expression from 3 to 28 days. We showed previously that HDAd expresses a transgene stably in EC from 28 days through at least 48 weeks;11 therefore, it now appears that HDAd transduction of EC in vivo can stably express transgenes from 3 days onward. Accordingly, no further efforts are required either to prevent early loss of HDAd genomes or early loss of transgene expression. We also discovered that elimination of the IL-10 3′-UTR dramatically increases HDAd expression of IL-10. Lack of atheroprotective effects of IL-10 in a previous study were likely due to low expression levels,15 and it will be valuable to repeat this study with the higher-expressing IL-10 vector. Finally, we identified a 51 bp segment of the apo A-I 3′-UTR that likely plays a major role in mRNA stability in EC.
There are limitations to our study. These include variations in vector structure that are not perfectly controlled in individual experiments. For example, the 4XETE vectors all contain the oPRE, whereas the CMV vectors do not. Although it is possible that differences between 4XETE and CMV promoter performance (e.g., data in Fig. 4b showing lower and more stable expression from 4XETE) could be attributed to the oPRE, this seems unlikely because the oPRE increases transgene expression in this system and in others;14,49 whereas, in this experiment (Fig. 4b) the 4XETE/oPRE-containing construct expresses at levels equal to or far below those obtained with the CMV promoter. Another limitation is that some of our experiments were performed in vitro, and may not predict in vivo results. However, we tested rabbit codon-optimization approaches in cultured rabbit cells (fibroblasts), in which rabbit-specific codon optimization should be effective. We also initially tested the 3′-UTR deletion approach in vitro. In this case, our positive results in vitro results were replicated—even more impressively—in vivo. Our use of cultured bovine EC rather than rabbit EC could also yield in vitro results that are not applicable to our in vivo model. This concern is somewhat mitigated by our finding that rabbit IL-10-containing conditioned medium regulated expression of the 4XETE-containing vector present in bovine EC, showing that bovine EC likely respond to rabbit IL-10. Unfortunately, we are no longer able to obtain rabbit EC.
In conclusion, when compared to previous work that addresses either transcriptional control in HDAd or optimization of transgene expression in EC in vivo,43,50 our results reveal new factors that influence the level and time course of transgene expression after in vivo transduction of EC with HDAd. These factors include susceptibility of the vector promoter to acute upregulation as well as feedback effects of transgene products (such as IL-10) on transgene expression. The CMV promoter is suitable for applications in which an initial pulse of transgene expression is desirable. At 28 days, the CMV and 4XETE promoters perform similarly. We did not compare their performance at later time points, although both appear to express stably beyond 4 weeks.11,15 Because the CMV promoter is not cell-type specific and is susceptible to downregulation in vivo, future efforts to develop HDAd for treatment of chronic vascular diseases such as atherosclerosis will be focused on increasing expression from stably expressing mammalian-derived endothelial-selective promoters such as 4XETE.
Supplementary Material
Acknowledgments
This work was supported in part by grant HL114541 from the NIH and by the John L. Locke, Jr. Charitable Trust. We thank AdVec, Inc. for permission to use the HDAd reagents, Thaïs Guimaraes and Meena Sethuraman for technical assistance, and Julia Feyk for administrative assistance.
Author Disclosure
No competing financial interests exist.
References
- 1.De Caterina R, Libby P, eds. Endothelial Dysfunctions and Vascular Disease. Malden, MA: Blackwell, 2007 [Google Scholar]
- 2.Gimbrone MA, Jr., Garcia-Cardena G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res 2016;118:620–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White SJ, Nicklin SA, Buning H, et al. Targeted gene delivery to vascular tissue in vivo by tropism-modified adeno-associated virus vectors. Circulation 2004;109:513–519 [DOI] [PubMed] [Google Scholar]
- 4.Anliker B, Abel T, Kneissl S, et al. Specific gene transfer to neurons, endothelial cells and hematopoietic progenitors with lentiviral vectors. Nat Methods 2010;7:929–935 [DOI] [PubMed] [Google Scholar]
- 5.Abel T, El Filali E, Waern J, et al. Specific gene delivery to liver sinusoidal and artery endothelial cells. Blood 2013;122:2030–2038 [DOI] [PubMed] [Google Scholar]
- 6.Lipinski DM, Reid CA, Boye SL, et al. Systemic vascular transduction by capsid mutant adeno-associated virus after intravenous injection. Hum Gene Ther 2015;26:767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemarchand P, Jaffe HA, Danel C, et al. Adenovirus-mediated transfer of a recombinant human a1-antitrypsin cDNA to human endothelial cells. Proc Natl Acad Sci U S A 1992;89:6482–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulick AH, Dong G, Newman KD, Virmani R, Dichek DA. Endothelium-specific in vivo gene transfer. Circ Res 1995;77:475–485 [DOI] [PubMed] [Google Scholar]
- 9.Vassalli G, Agah R, Qiao R, Aguilar C, Dichek DA. A mouse model of arterial gene transfer. Antigen-specific immunity is a minor determinant of the early loss of adenovirus-mediated transgene expression. Circ Res 1999;85:e25–e32 [PubMed] [Google Scholar]
- 10.Parks RJ, Chen L, Anton M, Sankar U, Rudnicki MA, Graham FL. A helper-dependent adenovirus vector system: Removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci U S A 1996;93:13565–13570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flynn R, Qian K, Tang C, et al. Expression of apolipoprotein A-I in rabbit carotid endothelium protects against atherosclerosis. Mol Ther 2011;19:1833–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen S, Graf S, Massey PG, Dichek DA. Improved vascular gene transfer with a helper-dependent adenoviral vector. Circulation 2004;110:1484–1491 [DOI] [PubMed] [Google Scholar]
- 13.Du L, Zhang J, Clowes AW, Dichek DA. Efficient gene transfer and durable transgene expression in grafted rabbit veins. Hum Gene Ther 2015;26:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dronadula N, Du L, Flynn R, et al. Construction of a novel expression cassette for increasing transgene expression in vivo in endothelial cells of large blood vessels. Gene Ther 2011;18:501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du L, Dronadula N, Tanaka S, Dichek DA. Helper-dependent adenoviral vector achieves prolonged, stable expression of interleukin-10 in rabbit carotid arteries but does not limit early atherogenesis. Hum Gene Ther 2011;22:959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Anton M, Graham FL. Production and characterization of human 293 cell lines expressing the site-specific recombinase Cre. Somat Cell Mol Genet 1996;22:477–488 [DOI] [PubMed] [Google Scholar]
- 17.Flynn R, Buckler JM, Tang C, Kim F, Dichek D. Helper-dependent adenoviral vectors are superior in vitro to first-generation vectors for endothelial cell-targeted gene therapy. Mol Ther 2010;18:2121–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman KD, Dunn PF, Owens JW, et al. Adenovirus-mediated gene transfer into normal rabbit arteries results in prolonged vascular cell activation, inflammation, and neointimal hyperplasia. J Clin Invest 1995;96:2955–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider DB, Vassalli G, Wen S, et al. Expression of Fas ligand in arteries of hypercholesterolemic rabbits accelerates atherosclerotic lesion formation. Arterioscler Thromb Vasc Biol 2000;20:298–308 [DOI] [PubMed] [Google Scholar]
- 20.Jiang B, Qian K, Du L, Luttrell I, Chitaley K, Dichek DA. Helper-dependent adenovirus is superior to first-generation adenovirus for expressing transgenes in atherosclerosis-prone arteries. Arterioscler Thromb Vasc Biol 2011;31:1317–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 2004;313:856–862 [DOI] [PubMed] [Google Scholar]
- 22.Kozera B, Rapacz M. Reference genes in real-time PCR. J Appl Genet 2013;54:391–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Pennell ML, Pearl DK, Knobloch TJ, Fernandez S, Weghorst CM. The choice of reference gene affects statistical efficiency in quantitative PCR data analysis. Biotechniques 2013;55:207–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fath S, Bauer AP, Liss M, et al. Multiparameter RNA and codon optimization: a standardized tool to assess and enhance autologous mammalian gene expression. PLoS One 2011;6:e17596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp PM, Li WH. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 1987;15:1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci 2012;69:3613–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Vries JE. Immunosuppressive and anti-inflammatory properties of interleukin 10. Ann Med 1995;27:537–541 [DOI] [PubMed] [Google Scholar]
- 28.Chen D, Ding Y, Zhang N, et al. Viral IL-10 gene transfer inhibits the expression of multiple chemokine and chemokine receptor genes induced by inflammatory or adaptive immune stimuli. Am J Transplant 2003;3:1538–1549 [DOI] [PubMed] [Google Scholar]
- 29.Papadakis ED, Nicklin SA, Baker AH, White SJ. Promoters and control elements: designing expression cassettes for gene therapy. Curr Gene Ther 2004;4:89–113 [DOI] [PubMed] [Google Scholar]
- 30.Qin L, Ding Y, Pahud DR, Chang E, Imperiale MJ, Bromberg JS. Promoter attenuation in gene therapy: interferon-gamma and tumor necrosis factor-alpha inhibit transgene expression. Hum Gene Ther 1997;8:2019–2029 [DOI] [PubMed] [Google Scholar]
- 31.Loser P, Jennings GS, Strauss M, Sandig V. Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NFkappaB. J Virol 1998;72:180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritter T, Brandt C, Prosch S, et al. Stimulatory and inhibitory action of cytokines on the regulation of hCMV-IE promoter activity in human endothelial cells. Cytokine 2000;12:1163–1170 [DOI] [PubMed] [Google Scholar]
- 33.Quax TE, Claassens NJ, Soll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol Cell 2015;59:149–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foster H, Sharp PS, Athanasopoulos T, et al. Codon and mRNA sequence optimization of microdystrophin transgenes improves expression and physiological outcome in dystrophic mdx mice following AAV2/8 gene transfer. Mol Ther 2008;16(11):1825–1832 [DOI] [PubMed] [Google Scholar]
- 35.Valencik ML, McDonald JA. Codon optimization markedly improves doxycycline regulated gene expression in the mouse heart. Transgenic Res 2001;10:269–275 [DOI] [PubMed] [Google Scholar]
- 36.Gustafsson C, Minshull J, Govindarajan S, Ness J, Villalobos A, Welch M. Engineering genes for predictable protein expression. Protein Expr Purif 2012;83:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maertens B, Spriestersbach A, von Groll U, et al. Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli. Protein Sci 2010;19:1312–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welch M, Villalobos A, Gustafsson C, Minshull J. You're one in a googol: optimizing genes for protein expression. J R Soc Interface 2009;6 Suppl 4:S467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paulding WR, Czyzyk-Krzeska MF. Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rich sequence in the 3′-untranslated region. J Biol Chem 1999;274:2532–2538 [DOI] [PubMed] [Google Scholar]
- 40.Miao CH, Ohashi K, Patijn GA, et al. Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro. Mol Ther 2000;1:522–532 [DOI] [PubMed] [Google Scholar]
- 41.Zhao W, Pollack JL, Blagev DP, Zaitlen N, McManus MT, Erle DJ. Massively parallel functional annotation of 3′ untranslated regions. Nat Biotechnol 2014;32:387–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han X, Kitamoto S, Lian Q, Boisvert WA. Interleukin-10 facilitates both cholesterol uptake and efflux in macrophages. J Biol Chem 2009;284:32950–32958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schiedner G, Hertel S, Johnston M, Biermann V, Dries V, Kochanek S. Variables affecting in vivo performance of high-capacity adenovirus vectors. J Virol 2002;76:1600–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Golden CL, Nick HS, Visner GA. Thrombin regulation of endothelin-1 gene in isolated human pulmonary endothelial cells. Am J Physiol 1998;274:L854–863 [DOI] [PubMed] [Google Scholar]
- 45.Quehenberger P, Bierhaus A, Fasching P, et al. Endothelin 1 transcription is controlled by nuclear factor-kappaB in AGE-stimulated cultured endothelial cells. Diabetes 2000;49:1561–1570 [DOI] [PubMed] [Google Scholar]
- 46.Douthwaite JA, Lees DM, Corder R. A role for increased mRNA stability in the induction of endothelin-1 synthesis by lipopolysaccharide. Biochem Pharmacol 2003;66:589–594 [DOI] [PubMed] [Google Scholar]
- 47.Ross PJ, Kennedy MA, Christou C, Risco Quiroz M, Poulin KL, Parks RJ. Assembly of helper-dependent adenovirus DNA into chromatin promotes efficient gene expression. J Virol 2011;85:3950–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong CM, McFall ER, Burns JK, Parks RJ. The role of chromatin in adenoviral vector function. Viruses 2013;5:1500–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schambach A, Bohne J, Baum C, et al. Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther 2006;13:641–645 [DOI] [PubMed] [Google Scholar]
- 50.White SJ, Papadakis ED, Rogers CA, Johnson JL, Biessen EA, Newby AC. In vitro and in vivo analysis of expression cassettes designed for vascular gene transfer. Gene Ther 2008;15:340–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.