Figure 5.
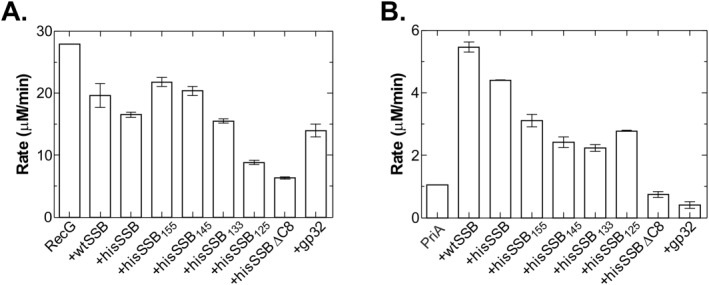
SSB linker mutants affect RecG and PriA differently. ATPase assays were carried out as described in Materials and methods. For RecG, the DNA substrate was M13 ssDNA and for PriA, it was ϕx174 ssDNA. For each helicase, DNA substrate (10 μM nucleotides) was incubated separately with each single strand binding protein at 37°C for 2 min, prior to the addition of RecG (100 nM, final) or separately, PriA (10 nM, final) to initiate reactions. In RecG assays, SSB proteins were present at 125 nM tetramer and gp32 at 500 nM monomer. For PriA, SSB proteins were present at 200 nM tetramer and gp32 was 800 nM monomer. (A) Linker deletion mutants inhibit RecG more than wtSSB. (B) Linker mutants are not effective stimulators of PriA. The analysis in each panel represents two assays done on the same day. Error bars represent the standard deviation.
