Figure 1.
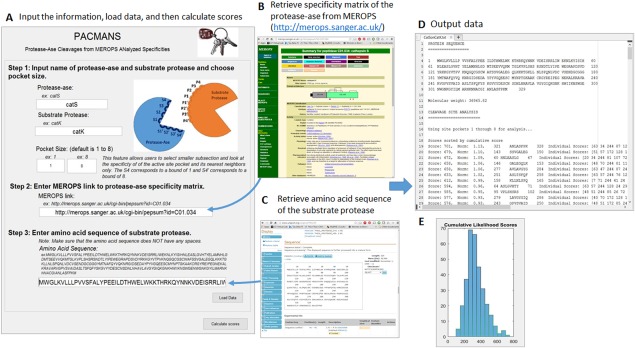
Using PACMANS to score locations of hydrolysis of the substrate protease by the protease‐ase. Input the name of hypothesized protease‐ase and desired substrate protease, specificity matrix for the protease‐ase, and amino acid sequence of desired substrate protease. The specificity matrix quantifies the likelihood that a given amino acid in positions P4 through P4′ will result in a peptide bond cleavage between P1 and P1′ in the enzyme's active site. Using the protease‐ase's specificity matrix and another protease's amino acid sequence as the substrate protease, the potential locations of cleavage were scored by the sum of the individual scores that a given residue has been reported to occur in each subsite of the active site pocket. These putative site of cleavage by protease‐ase were then sorted by score and given to users in text file along with a histogram of all the scores.
