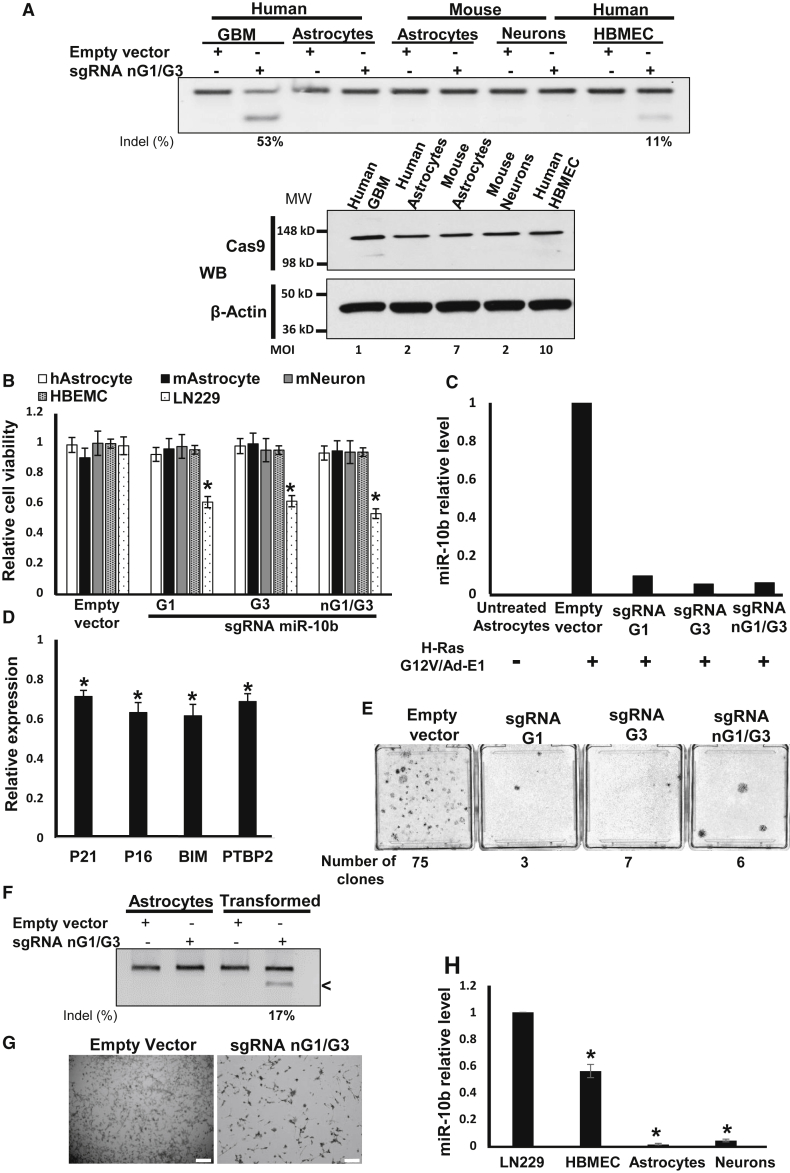
Figure 5.
Lentivirus-Mediated miR-10b Gene Editing Abolishes Neoplastic Transformation of Oncogene-Induced Astrocytes
(A) Transductions of human and mouse primary astrocytes and neurons with miR-10b-editing lentivirus at the MOI levels that led to similar levels of Cas9 expression, as assessed by western blot with Cas9 antibody (low panel), does not cause miR-10b gene editing. In these conditions, 100% of glioma LN229 cells were Cas9 positive. Human brain microvascular endothelial cells (HBMECs) were edited in the miR-10b gene by high-titer virus with low efficiency (11% versus 53% in glioma cells, at a 10-fold higher viral titer). The relative MOI required for similar Cas9 expression in these cells is indicated. (B) miR-10b gene editing reduces the viability of glioma cells, but not human and mouse primary astrocytes, neurons, and HBMECs, as determined by WST1 assays 48 hr post-transduction. Transduction conditions and MOI match those used in (A). n = 6, *p < 0.001, Student’s t test. (C) miR-10b levels in mouse primary astrocytes induced for transformation by H-RasG12V/Ad-E1 and subsequently transduced with miR-10b-editing vectors for 2 weeks, as determined by qRT-PCR and normalized to the geometrical mean of unaffected miR-99a, miR-125a, and miR-148a. (D) Transformed primary astrocytes exhibit reduced levels of miR-10b targets p21, p16, BIM, and PTBP2, relative to the corresponding naive cultures. qRT-PCR data were normalized to the geometrical mean of three unaffected genes (GAPDH, 18S rRNA, and SERAC1). Error bars depict SEM, n = 3, *p < 0.05, Student’s t test. (E) miR-10b editing reduces the number of transformed colonies. Crystal violet staining and quantification of the colonies are shown 2 weeks after infections with miR-10b-editing vectors. (F) Transformed, miR-10b-expressing mouse astrocytes become editable in the miR-10b locus. (G) miR-10b editing of transformed astrocytes induces cell death, similar to the effect on glioma cell lines. Scale bar, 20 μm. (H) Relative miR-10b levels in glioma and various brain-derived cell types were assessed by qRT-PCR analysis, and the data were normalized to the geometrical mean of unaffected miR-99a, miR-125a, and miR-148a. n = 6, *p < 0.001, Student’s t test.