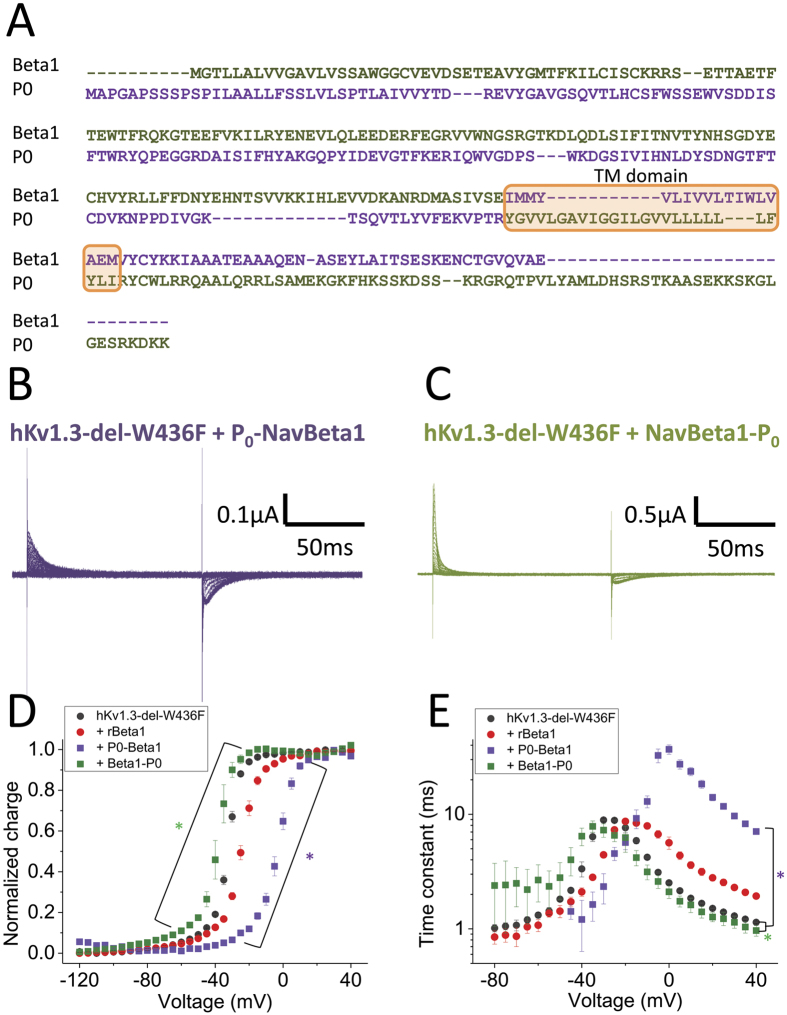
Figure 4. The effect of chimeras of rNavBeta1 and P0 on hKv1.3 gating current.
(A) Amino acid sequence alignment between rNavBeta1 and myelin protein zero (P0), a protein with high homology to rNavBeta1. Transmembrane segment is highlighted by orange rectangle. The sequence for NavBeta1-P0 chimera is indicated in green and that for P0-NavBeta1 in purple. (B,C) Representative gating currents from hKv1.3-del-W436F co-injected with P0-NavBeta1 (B) or with NavBeta1-P0 (C). (D,E) The Q-V relationship (D) and Tau-V relationship (E) for hKv1.3-del-W436F alone (black circles, n = 8), co-injected with rNavBeta1 (red circles, n = 5), co-injected with P0-NavBeta1 (purple squares, n = 5) or co-injected with NavBeta1-P0 (green squares, n = 3). Error bars indicate SEM. Green stars (*) indicate statistical significance (p-value < 0.05) of the difference between hKv1.3-del-W436F alone (black circles) and with NavBeta1-P0 (green squares) from −95 mV to −10 mV in Q-V (D), and at −15, −10, +5, +20 and +40 mV in Tau-V (E). Purple stars (*) indicate statistical significance (p-value < 0.05) of the difference between hKv1.3-del-W436F alone (black circles) and with P0-NavBeta1 (purple squares) from −70 mV to +20 mV in Q-V (D), and from −40 mV to +40 mV except at −20 mV in Tau-V (E).