Figure 1.
Generation of an In Vitro Model of LCA10 Using CRISPR/Cas9
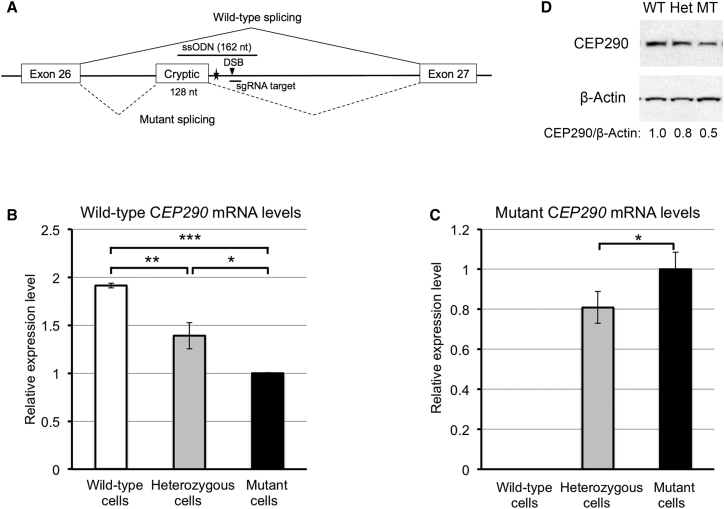
(A) A schematic diagram showing the IVS26 mutation (filled star) in the CEP290 gene and the locations of the sgRNA target (antisense strand) and ssODN (sense strand) used for introducing the IVS26 mutation. The DSB site (filled triangle) induced by SpCas9 was located 15 bp downstream of the IVS26 mutation. The splicing patterns for wild-type and mutant are represented by solid and dashed lines, respectively. (B and C) Basal levels of wild-type CEP290 mRNA (B) and mutant CEP290 mRNA (C) in the wild-type (white bar), heterozygous (gray bars), and mutant 293FT cells (black bars), as determined by qRT-PCR. CEP290 mRNA levels were normalized to the levels of Cyclophilin A (PPIA) mRNA. The data are presented as the means ± SD (n = 3). Comparisons were performed using one-way ANOVA followed by the Tukey honest significant difference (HSD) post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001. (D) Immunoblot analysis of lysates prepared from the wild-type (WT), heterozygous (Het), and mutant 293FT cells (MT). The membrane was probed for CEP290 protein (top) and β-actin (bottom; as a loading control). The ratio of CEP290/β-actin is shown at the bottom, with the ratio for the wild-type cells set to 1.0.