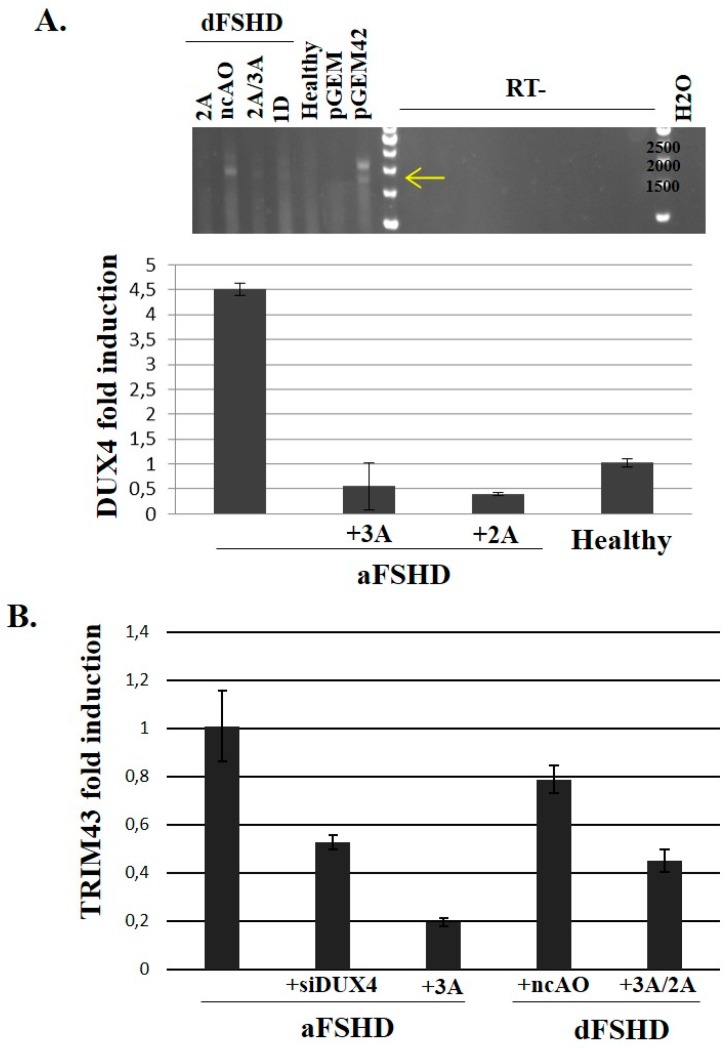
Figure 5.
Evaluation of DUX4-AO efficiency on endogenous DUX4 and target mRNA expression in FSHD primary myotubes. 105 primary myoblasts (control, aFSHD and dFSHD) were seeded in 35 mm culture dishes. The next day, cells were transfected with either the negative control AO (nc-AO, 600 nM) or AOs pLAM2A(−7 + 18) and pLAM3A(−12 + 13) either alone (lanes 2A or 3A) or in a cocktail (lane 2A/3A) and pLAM1D(+7 − 18) (lane 1D) at previously determined optimal concentrations. Differentiation was induced 4 h after transfection and the cells were harvested 72 h later. (A) Top: Total RNAs of dFSHD myotubes were extracted and submitted to RT-PCR with primers we had previously shown to be specific of the DUX4 full length ORF [19]. The RT-PCR products were separated by electrophoresis on an agarose gel and stained with ethidium bromide. The controls were total RNAs of C2C12 cells transfected with the pGEM plasmid either without insert (negative control) or with a genomic fragment containing 2 D4Z4 units [18] (pGEM42). The experiment was done in the presence (RT+) or absence (RT-) of retrotranscriptase to demonstrate the products did not result from amplification of contaminating genomic DNA. Bottom: Total RNAs of aFSHD myotubes were extracted and submitted to reverse transcription (RT) and amplification by qPCR with DUX4-specific primers. The relative abundance was calculated using Ribosomal Protein Lateral Stalk Subunit P0 (RPLPO) as a reference for cDNA input and following M. Pfaffl’s guidelines [68,69]. The data are presented as fold change in DUX4 mRNA abundance with or without AO treatment; (B) Fold change in TRIM43 mRNA abundance after treatment with an AO or siRNA against DUX4 mRNA. The RT-qPCR was performed as described [67].