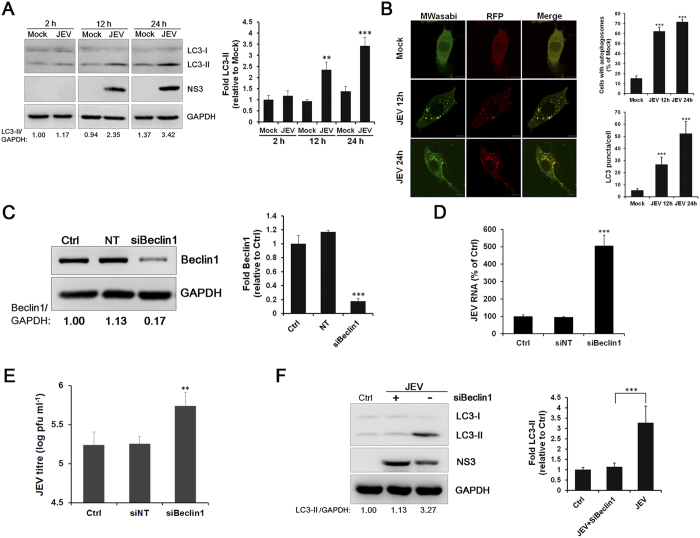
Figure 5. JEV replication is negatively regulated by autophagy in SK-N-SH cells.
(A) SK-N-SH cells were infected with JEV (MOI = 1.0), and the cells were collected at 2 h, 12 h, and 24 h p.i. The protein levels of LC3-I, LC3-II and NS3 were evaluated by Western blot analysis. GAPDH served as a control for equal sample loading. The relative quantification of the detected signal was analysed using ImageJ software and normalized to GAPDH. (B) Representative fluorescent images and statistical results of punctate mTagRFP-mWasabi-LC3. Cells with autophagosomes were chosen from a pool of at least 10 images. Scale bar is 10 μm. (C) SK-N-SH cells were transfected with Beclin1 siRNA (50 nM) or the NT siRNA (50 nM). The level of Beclin1 protein was detected by Western blot. GAPDH was detected as a loading control. The relative quantification of the detected signal was analysed using ImageJ software and normalized to GAPDH. (D and E) The virus RNA was analysed by RT-qPCR (D), and the titres of the culture supernatants from Beclin1-depleted cells were determined by plaque forming unit assays (E). (F) SK-N-SH cells transfected with Beclin1 siRNA (50 nM) or the NT siRNA (50 nM) were infected with JEV (MOI = 1.0). The levels of LC3-I, LC3-II and NS3 were evaluated by Western blot analysis. GAPDH was detected as a loading control. The data are shown as the mean ± SD of three independent experiments. **, P < 0.01; ***, P < 0.001.