Fig. 2.
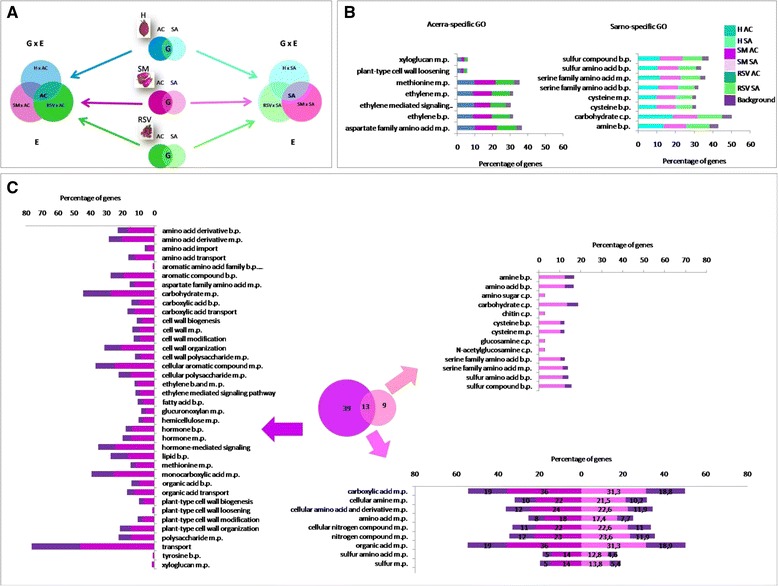
Gene ontology enrichment analysis. a Scheme for classifying over-represented gene classes. For each genotype Acerra-specific (Ac) and Sarno-specific (Sa) enriched GO terms were identified. Enriched GO terms common to both environments (G) in each genotype (H, SM and RSV) were also identified. By crossing enriched GO terms in Acerra from all three genotypes, Acerra-specific and Acerra × Genotype interactions were identified. The same scheme was used to identify Sarno-specific enriched GO terms as well as Sarno × Genotype interactions. b Environment-specific enriched GO categories. Left) Acerra-specific enriched GO terms. Right) Sarno-specific enriched GO terms. c San Marzano GO Enrichment Analysis. The Venn diagram shows common and specific enriched GO terms. Bar plots reflect the percentage of genes in the enriched categories of the San Marzano Acerra (left), Sarno (right) and common (below), as well as the percentage of genes belonging to the same categories in tomato genome. Common enriched GO categories are reported for both environments because some categories, although enriched in both conditions, have a different percentage of genes. m.p. = metabolic process, b.p. = biological process, c.p. = catabolic process
