Fig. 3.
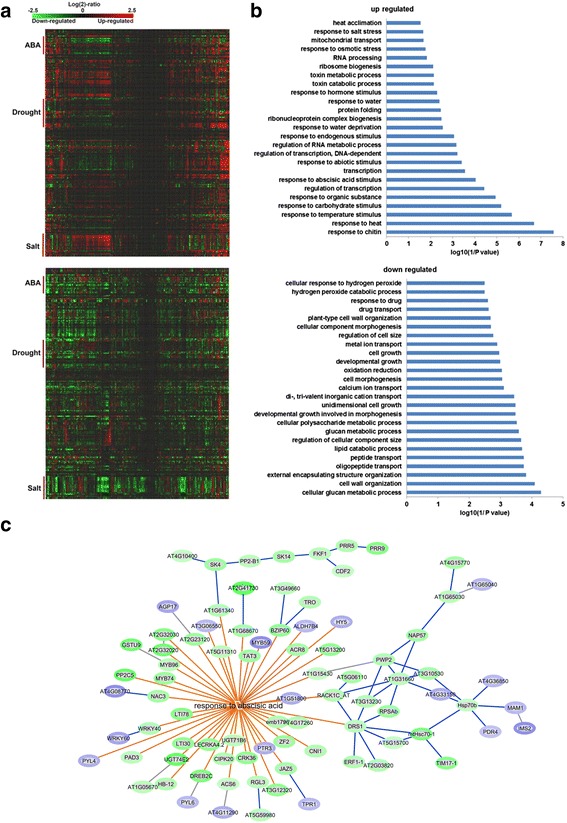
Gene expression changed by GEX1A corresponding to stress responses. a, upper panel, a heatmap was generated by mapping 400 randomly chosen upregulated genes in GEX1A treatment to the microarray database using Genevestigator. The heatmap indicates that a great number of these genes are upregulated (red) by ABA, drought and salt stress. Bottom panel: a heatmap was generated by mapping 400 randomly chosen down-regulated genes in the GEX1A treatment to the microarray database using Genevestigator. The heatmap indicates that a great number of these genes are downregulated (green) by ABA, drought, and salt stress. b, Functional categorization of regulated genes. Functional categorization (biological process) of up/downregulated genes in GEX1A treatment. Top 25 enriched pathways are shown. c, Differentially expressed genes in GEX1A treatment were mapped onto the response-to-abscisic-acid pathway. The analysis was performed using the Exploratory Gene Association Networks (EGAN) software tool. Orange lines show the participation of the genes in abscisic acid-activated signaling pathways and blue lines show known interactions between the genes connected. Green ovals represent upregulated genes and blue ovals represent downregulated genes
