Abstract
Cells in the body are physically confined by neighboring cells, tissues, and the extracellular matrix. Although physical confinement modulates intracellular signaling and the underlying mechanisms of cell migration, it is difficult to study in vivo. Furthermore, traditional two-dimensional cell migration assays do not recapitulate the complex topographies found in the body. Therefore, a number of experimental in vitro models that confine and impose forces on cells in well-defined microenvironments have been engineered. We describe the design and use of microfluidic microchannel devices, grooved substrates, micropatterned lines, vertical confinement devices, patterned hydrogels, and micropipette aspiration assays for studying cell responses to confinement. Use of these devices has enabled the delineation of changes in cytoskeletal reorganization, cell–substrate adhesions, intracellular signaling, nuclear shape, and gene expression that result from physical confinement. These assays and the physiologically relevant signaling pathways that have been elucidated are beginning to have a translational and clinical impact.
Keywords: cell confinement, microfluidics, micropatterning, cell migration, cytoskeletal organization, mechanosensing
1. INTRODUCTION
Cells in the body are confined by and respond to topography, be it imposed by other cells or by components of the extracellular matrix (ECM). The ECM consists of both randomly oriented fibers and more complex topographies that present two-dimensional (2D) or three-dimensional (3D) guidance cues to migrating cells (1). Cells display great plasticity in their migration mechanisms and modulate signaling pathways and intracellular cytoskeleton and adhesion organization in response to physical constraints (2–5). Therefore, identifying physical factors driving this adaptability and discovering methods to control, manipulate, promote, or stop migration over the full range of mechanisms available to cells are vital for understanding development and morphogenesis (6, 7), tuning the immune response (8, 9), engineering highly structured tissues (10), and combating cancer metastasis (1, 11). However, studying cell response to confinement in vivo is costly and low throughput. Therefore, engineers have devised models to recapitulate important aspects of the microenvironment so as to rapidly analyze the effect of well-defined physical inputs on cell behavior.
In this review, we discuss bioengineering models used to probe the mechanisms of confined cell migration and signaling. These models include polydimethylsiloxane (PDMS) microchannel devices, grooved substrates, microcontact printed substrates, vertically confining devices, patterned hydrogels, and micropipette aspiration assays. These assays span the range of guidance cues found in vivo, from tracks devoid of barriers to 3D scaffolds presenting the cells with many obstacles (1). After discussing the design and use of these devices, we describe changes in cytoskeletal architecture, cellular adhesions, cell signaling pathways, and gene expression that are induced by cell confinement. We conclude with several case studies demonstrating how exploiting cell behavior in confinement can be used to improve human health.
2. EXPERIMENTAL MODELS FOR STUDYING CELL BEHAVIOR IN CONFINEMENT
In 2D environments, epithelium-derived cells migrate in a cyclic fashion. Cellular adhesions anchor the cell body while new protrusions are generated, and as the cell moves forward, adhesions at the trailing edge release (12, 13). This migration mode depends on the formation of adhesion sites, actin-driven protrusion generation, and contractility in which myosin motors pull against actin fibers to create stress within the cell (12, 13). However, this traditional model of cell migration does not hold, at least in all cases, in more complex environments (4, 14–17). Cells in vivo migrate through confining 3D spaces encompassing both fibrillar ECM gels and preexisting migration tracks (1). To mimic the earmarks of the in vivo microenvironment, a series of devices have been engineered that impose various types of confinement on cells (Figure 1).
Figure 1.
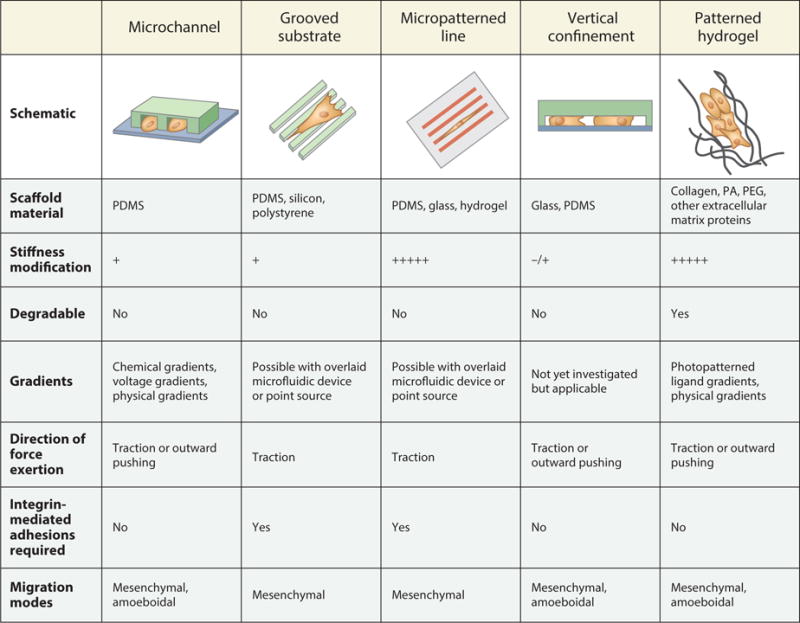
Schematics of engineered models of cell migration, along with their key physical, mechanical, and mechanistic characteristics. Microfluidic microchannel devices, grooved substrates, micropatterned lines, vertical confinement devices, and patterned hydrogels can be engineered to study cells’ response to confinement. Scaffolds for confined cell migration can be formed from various degradable and nondegradable materials [including polydimethylsiloxane (PDMS), polyacrylamide (PA), and polyethylene glycol (PEG)] with tunable stiffness. Additionally, these scaffolds can be combined with gradients to simultaneously impart multiple cues to migrating cells. Use of these assays has revealed microenvironment-dependent patterns of force exertion and requirements for integrin-mediated adhesions. Importantly, the physical and chemical properties of the model can drive cells to migrate via mesenchymal or amoeboidal mechanisms.
2.1. Polydimethylsiloxane Microfluidic Microchannel Devices
Many devices fabricated to study cell migration in 3D confinement are composed of PDMS, an elastomer that is optically transparent and through which oxygen can diffuse (18–20). PDMS devices for confined migration studies are typically produced by micromolding PDMS from a patterned silicon wafer (21). In these devices, microchannels arrayed adjacent to cell seeding areas allow for direct, real-time imaging of cell migration under confining conditions and in the absence of shear stress (22). Channels can be coated with a variety of ECM proteins (3, 23) and designed to explore both basic mechanisms of migration in confinement and cells’ response to external gradients. PDMS channels recapitulate aspects of “barrier-free” migration tracks in the lumen of vessels, in bone cavities, through clefts between muscle and nerve fibers, along bundled collagen tracks left by invading “leader” cells, and in perivascular and white matter tracks in the brain (1, 24–30), making them a versatile tool to study confined migration. These devices are illustrated schematically in Figure 1.
2.1.1. Straight polydimethylsiloxane microchannels
Microchannels fabricated for the study of cell migration during physical confinement are typically straight, shaftlike spaces ranging in cross-sectional area from ∼20 μm2 to greater than 1,000 μm2 (2–4, 14, 22, 23, 31–37). Early studies of microchannel migration examined the effects of varying channel cross-sectional areas on cell migration speed. Irimia & Toner (23) studied the spontaneous migration of tumor cells through 600-μm-long PDMS microchannels of varying dimensions (3 or 12 μm in height and ranging from 6 to 100 μm in width). The migration speed of MDA-MB-231 cells in 3-μm-tall channels peaked in 25-μm-wide channels and showed an overall biphasic response to cross-sectional area, whereas speed was not a function of channel width in the taller microchannels (23). Migration speed in microchannels is often greater than that observed in 2D environments, and fully confined cells migrate in a sliding motion, with small deviations in cell length (14, 37). In general, cells do not efficiently penetrate into smaller microfabricated pores (37). It is currently unknown whether vertical (for example, 3-μm-tall by 50-μm-wide channels) and lateral (for example, 50-μm-tall by 3-μm-wide channels) confinement equivalently affect cell migration within microchannels.
2.1.2. Chemical gradients
Chemotaxis is important in tumor dissemination (38) and the immune response (39). The lack of convective flow through microchannels enables formation of chemical gradients between source and sink reservoirs located at the microchannel inlets and outlets (22). Therefore, these assays are readily adaptable for chemotaxis studies. The presence of a gradient establishes a preferential direction for cell transmigration through microchannels (40). For example, Tong et al. (3) described a microfluidic device through which cells migrate up a fetal bovine serum (FBS) chemotactic stimulus through 10-μm-tall channels varying in width from 3 to 50 μm. The device showed differential migration of metastatic versus nonmetastatic cells; MDA-MB-231 cells were more likely to enter and exit microchannels than tumorigenic but nonmetastatic MCF-7 cells (3). This finding suggests that such devices could be used to screen for metastatic cells. Assays that rely on microchannel chemotactic migration could also be used in the presence of various pharmacological agents, allowing for screening of drug candidates (41, 42). A device recently constructed by Zhang et al. (43) contained 3,120 microchannel-incorporating microchambers and allowed for simultaneous screening of 9 pharmacological inhibitors (with multiple technical replicates and seeding densities for each compound) during migration in microchannels up an FBS gradient. This screen revealed a differential migration response of SUM-159/Taxol cells to different inhibitors and showed the promise of high-throughput microchannel migration assays for drug-screening applications (43).
2.1.3. Voltage gradients
In addition to chemical gradients, cells respond to electrical fields. Direct current electric fields (~0.5–5 V/cm) are present in vivo across endothelial and epithelial surfaces and play a role in wound healing and cancer metastasis (44–46). Huang et al. (46) tested the effects of combining confinement (20-μm-wide, 80-μm-tall PDMS channels coated with fibronectin) and an electric field applied between two Ag/AgCl electrodes on the motility of NIH 3T3 cells. Whereas NIH 3T3 fibroblasts aligned perpendicularly to the electric field and migrated toward the cathode when they were unconfined, they aligned and migrated along microchannels when confined, suggesting that topographical cues provide a stronger alignment cue than do electric fields (46). When no electric field was present, cell migration in the microchannels was significantly faster than that on 2D surfaces (46). In the presence of a 2.2 V/cm electric field, cells migrated along the channels and toward the cathode, with a persistence significantly greater than when in the absence of a voltage gradient (46).
2.1.4. Physical gradients and cell decision making
Modification of microchannel devices to incorporate more complex channel geometries allows for testing of cells’ response to anisotropic physical gradients or cues that cells encounter in the evolving tumor microenvironment (1). Cells migrating in PDMS microchannels generally continue on their path when encountering an obstacle, although the geometry of the microenvironment and the cell type influence the likelihood of repolarization. MDA-MB-231 breast tumor cells migrating in 15-μm-wide microchannels that constrict to a width of 3.3 μm go through a delay period prior to moving fully through the pores; the delay period increases as the length of the 3.3-μm-wide pore increases (33). In general, some fraction of cells encountering such a constriction reverses direction instead of moving through the pore; this fraction varies between 14% and 88%, depending on the cell type and geometry of the constriction (32). When the decision region is made more complex by incorporation of a 90° split to 10- or 3.3-μm-wide channels, more than 90% of the cells choose the wider channel, a trend that holds when actomyosin contractility is inhibited by application of blebbistatin (47). If the 3.3-μm-wide channel is placed in a path collinear to the original channel, the percentage of cells choosing the wider path drops to 68% and, interestingly, to only 33% when cells are treated with blebbistatin, suggesting that actomyosin contractility has a role in environmental sensing (47). Thus, the geometry of the microchannel interacts with cell signaling and mechanosensing pathways to modulate cell decision making. Cells migrating through narrow microchannel “mazes” may locally consume growth factors to create gradients that assist in finding the shortest migration path (48), but this strategy may not hold at bifurcations that are asymmetric or larger than the cell body (C.D. Paul & K. Konstantopoulos, unpublished data). In some cases, neutrophils migrating through bifurcating microchannels push water to sense the hydraulic resistance of each channel encountered and move into the channel with the lowest hydraulic resistance, in the presence or absence of a gradient (35). If a chemical gradient is present, neutrophils’ choice to migrate into the shorter channel may also be due to the presence of a steeper gradient in the shorter channel (49). Whether such a mechanosensitive strategy dependent on hydraulic resistance can be adopted by tumor cells, which migrate more slowly and rely on the permeation of water through the cell body for confined migration (4), is currently unknown.
2.2. Grooved Substrates
Micromachined and microimprinted substrates contain nanometer- to micrometer-scale topographies on otherwise planar substrates, typically in the form of parallel grooves (Figure 1). These substrates are used to model the micro- to nanotopography of ECM fibers that serve as directional cues for migrating cells (50, 51). Grooves can be directly machined on silicon substrates or molded on PDMS or polystyrene surfaces. The width, depth, and spacing of these grooves determine whether cells migrate between grooves (52), analogous to along one side wall of a very tall microchannel, or span grooves to integrate subcellular topographic cues (53–55). Cells plated on grooved substrates typically align and migrate along the long axis of the grooves (54, 56). When the groove spacing is less than the width of the cell, groove depth appears to play a larger role in causing cell polarization than the distance between grooves. For example, corneal epithelial cells more strongly align along 600-nm-deep versus 150-nm-deep grooves (53). Confluent monolayers of both epithelial and fibroblast cells also respond to substrate topography, migrating along 1-μm-wide, 150-nm-deep PDMS grooves spaced 1 μm apart with greater persistence than on flat surfaces (57). Interestingly, the topographical cues from the patterned region of the substrate were propagated onto a neighboring flat region of the device to cause oriented migration over a distance of approximately nine cell widths (57). Cell-to-cell propagation of these guidance cues was unchanged when cell–cell adhesion was disrupted with a function-blocking antibody to E-cadherin or when cell contractility was disrupted (57). Instead, propagation distances for aligned migration decreased when cell density was reduced, indicating that cell crowding instead of biochemical cell–cell interactions is required to transmit topographical cues through a cell sheet (57). Interestingly, on micropatterned lines (see the next section), disruption of cell–cell adhesions disrupts the propagation lengths of physical cues (58) and force transmission (59) in collective cell migration. In summary, the use of microgroove assays has demonstrated that cells respond to a wide range (nanometers to micrometers) of topographical cues.
2.3. Micropatterned Lines and Islands
Microcontact printing and its variants are used to laterally constrain cells on a 2D substrate (Figure 1). By controlling available spreading area, microcontact printing can be used to impose different shapes on either stationary (60) or migrating (61) cells. Microcontact printing is traditionally performed by “inking” an elastomeric stamp with a protein and transferring that protein to a substrate (62). Nonstamped regions are then backfilled with a polymer that is nonadhesive to cells (63). Recently, other methods have been developed. “Stamp-off ” approaches allow printing of adjacent, multiple-component patterns on the microscale (64). Optical ablation of a nonadhesive polymer followed by backfilling with a cell-adhesive protein enables fabrication of feature sizes down to ∼1 μm in width (65). Selective cross-linking of polymers through photolithography masks also can be used to generate islands of different stiffness or composition on a hydrogel background (66). Microcontact printed lines are often used to model ECM fibers, along which cells migrate in vivo (67, 68).
2.3.1. Micropatterned one-dimensional lines to study cell migration
Lateral confinement to constrain cell migration along a single axis is a useful tool for studying persistent cell migration, the effects of confinement on cell morphology, and intracellular signaling. The resulting migration has been termed one-dimensional (1D) migration, as cells are constrained to move back and forth in one direction when the width of the line is less than the width of the cell body. Doyle et al. (65) studied cell migration on 1D patterned fibronectin lines ranging in concentration from 0.5 to 1,000 μg/mL. Cells constrained to linear migration paths moved with significantly greater speed than cells in two dimensions, and speed was independent of ligand density (65). Uniaxial migration was lost if the ECM line width exceeded 5 μm, and migration speed decreased (65). On very wide lines, the speed of fibroblasts migrating collectively decreased as the cells’ distance from the edge of the line increased (69).
Microcontact printing techniques have also been used to show (a) that the Golgi apparatus and the microtubule-organizing center localize to the rear of the cells during persistent epithelial cell migration, with repositioning to the new trailing edge upon a change in cell direction (70), and (b) that the migration speed of MTLn3E tumor cells increases on 1D lines when the cells are cultured with bone marrow–derived macrophages (67). [Notably, rapid and directional tumor cell migration often occurs in vivo upon coupling of tumor cells and macrophages (67).] Furthermore, these assays provide lateral confinement to impose cell shape and enable studies of how the cytoskeleton and its links to the nucleus regulate nuclear shape (71), thereby allowing investigation into the relationship among nuclear shape, nuclear translocation, and cell migration (72). 1D migration assays have recently been used to demonstrate an exponential coupling between cell speed and persistence, in which persistence time increased as the actin retrograde flow rate increased (73). Microcontact printing migration assays can be further integrated with traction force microscopy to probe cell force exertion during lateral confinement (74). For example, NIH 3T3 fibroblasts exert lower traction stresses on 10-μm-wide printed lines than on 50-μm-wide printed lines (74). Note that although 1D migration on microprinted lines relies on cell adhesion to the substrate, alternatives to adhesion-based migration may be available in other microenvironments (15, 36).
2.3.2. Microcontact printed islands
The effects of confined geometry on cell polarization have been extensively studied through the use of microcontact printed “islands” on which cells adhere. Théry et al. (75) used microcontact printing to show that distinct directions of cell polarity could be established in cells with the same overall shape but differing underlying patterns of adhesion. Polarization is likely an initial cue for cell migration. When 3T3 fibroblasts are confined and forced to assume elongated morphologies via microcontact printing, cells migrate in the direction defined by the long axis of the cell when released (76). Changes in cell spreading area driven by the size of microcontact printed islands can also direct mesenchymal stem cell differentiation; human mesenchymal stem cells (hMSCs) differentiate to osteoblasts when plated on large printed islands but become adipocytes when seeded on small printed regions (77). Furthermore, microcontact printing can be extended to the printing of patterns on polyacrylamide (PA) gels (78) or micropost arrays (79) to study the effects of confinement on traction force exertion. In these assays, the traction forces exerted by a cell are related to either the displacement of fluorescent beads embedded in the gel (80) or the deflection of narrow pillars (81) on which the cells are seeded. Patterning of an ECM protein in a defined size and shape on these substrates enables decoupling of the effects of cell shape and substrate rigidity on force exertion.
2.3.3. Micropatterned stiffness islands
Microprinting and micropatterning have also been employed to define rigid islands (consisting of a cross-linked photoresist) in various configurations on a nonadhesive PA hydrogel of a given stiffness (66, 82). NIH 3T3 fibroblasts bridge gaps between adhesive regions on stiff (13-kPa) but not soft (0.8-kPa) gels; adhesions matured when the micropatterned island was not deflected toward the spreading cell (66). Confinement itself decreased the focal adhesion area (66), as has also been observed in narrow microchannels (14) and on microcontact printed lines (65). Cell spreading across rigid substrates depended on Rac1 and Arp 2/3, and the probing rate decreased upon inhibition of Cdc42 or formin homology 2 domain (66). Myosin II–mediated contractility was involved in both rigidity sensing (as determined by pulling on nascent adhesions to test the stiffness of the underlying substrate) and protrusion retraction (66). Indeed, when myosin II was inhibited by blebbistatin treatment, cells were not sensitive to rigidity and spread to occupy islands on soft substrates (66).
2.4. Vertical Confinement
As an alternative to inducing cells to migrate into confining microchannels or seeding cells on microcontact printed lines or grooved substrates, compressive forces can be dynamically and externally applied on cells via vertical confinement. Vertical confinement can simulate migration spaces observed between opposing surfaces in the body, such as in the space between peritoneal layers covering organs (1), and can induce blebbing migration observed in vivo through the peritumoral ECM (15, 83). Vertical confinement assays made possible by microfabrication techniques enable cell confinement to heights down to 3 μm, with an accuracy of 100 nm over large areas (square centimeters) (Figure 1) (84). In this technique, a pillared PDMS array is lowered over a bed of cells, and cells dispersed between the pillars are vertically confined to a height defined by the height of the pillars (84). This is a very high throughput technique if true 3D confinement is deemed unnecessary and vertical or so-called sandwich confinement is sufficient, and it is readily adapted to multiwall plates (84). Application of this technique has demonstrated how confinement can lead to rupture of the nuclear lamina and modulate gene expression pathways (85) and how mesenchymal cells can spontaneously switch to a rapid amoeboid migration mode when vertically confined (15). Aspects of vertical confinement assays can be recapitulated using compliant gels overlaid on cells or different materials on the basal and apical surfaces of cells (86), but this method sacrifices precise control of cell height.
2.5. Patterned Gels
Recent research has extended confinement assays to orthogonally probe the effects of substrate stiffness, ligand density, and microchannel size through patterning of mechanically tunable hydrogels (Figure 1). These studies are important complements to traditional 3D gel migration studies because they assay migration through defined microtracks, some of which are nondegradable. Migration tracks patterned in hydrogels mimic similar in vivo physiologies as PDMS microchannels but have more physiologically relevant stiffnesses, with the caveat that they are more difficult to fabricate. Both optical and micromolding techniques make fabrication of such structures possible.
2.5.1. Variable pore sizes via controlled polymerization
3D hydrogels with relatively homogeneous distribution of pores have been extensively used to model cell migration through fibrillar ECM. For example, both loosely and densely packed collagen matrices exist in the dermis and in mammary and interstitial tissues (87). One can vary pore sizes and fiber alignment in collagen gels by controlling polymerization conditions or using collagen stock with varying telopeptide content. For example, polymerizing rat tail collagen (1.7 mg/mL) at 9°C generates heterogeneous pore sizes (with a median pore cross section of 30 μm2), whereas polymerization at 37°C results in matrices with a median pore size of only 5 μm2 (88). Use of bovine dermal collagen with reduced telopeptide content results in approximately fivefold-larger pore sizes and a reduced matrix elastic modulus at the same overall collagen concentration (88). These techniques generate large pores that cells can navigate independently of matrix metalloproteinase (MMP) activity (88, 89). However, the dynamic range of pore sizes in 3D collagen gels is narrow, and the pores created are still often smaller than pores found in vivo (87). Cell-derived matrices (CDMs) demonstrate local orientation of fibers left by the cells depositing the matrix (90), but the spatial organization of the fibers is not controlled. CDMs are linearly elastic, and their use has been important in demonstrating pressure-based lobopodial migration (16, 17). However, because these techniques do not allow for control of track length and width, other methods have been developed.
2.5.2. Photopatterned gels
Photopatterning relies on the use of light to change or remove defined regions of a hydrogel. Controlling the delivery of light to hydrogels by using photolithography masks or focused laser light allows exquisite spatial and temporal control of light-based reactions, with ∼1-μm resolution in the x–y plane and ∼3–5-μm resolution in the z plane (91, 92). For example, ECM proteins and growth factors can be patterned within a 3D matrix to direct cell migration (93). Additionally, photodegradation reactions can be used either to degrade portions of the surface of a gel, resulting in microwells (94) or grooves (95), or to ablate regions within a collagen gel, leading to 3D tracks within the matrix that are bordered by degradable collagen (96). Such a scheme was used to study the invasion of multicellular spheroids of mouse mammary tumor cells embedded within dense fibrillar collagen (96). These cells invaded tracks with cross-sectional areas of 9, 25, 100, 400, and 900 μm2 in the absence of MMP activity and enlarged narrow tracks over time by an outward pushing mechanism. Orthogonal techniques for controlling ligand concentration and microtrack geometry enable “competition” assays to be performed. For example, 3T3 fibroblasts migrate through microchannels created around a fibrin clot, and when presented with a choice between channels functionalized with RGD peptide and those without, cells migrate only into channels with the peptide (91). Such approaches can also be used to temporally and spatially release cells or groups of cells from certain regions of a 3D gel for subpopulation sampling (91).
2.5.3. Micromolded collagen and polyacrylamide hydrogels
Polymers other than PDMS can be used to mold from a patterned master. This process enables orthogonal control of substrate stiffness and microchannel geometry (97) and can be performed with either degradable (31, 98) or nondegradable (97) polymers. Microtracks (10 μm wide, 20 μm tall, and 300 μm long) formed in collagen polymerized against a PDMS mold can be fabricated in 1.5, 2, 3, and 5 mg/mL collagen type I (31). The presence of microtracks increases the migration speed of MCF10A and MDA-MB-231 cells compared with unpatterned 3 mg/mL collagen gels (31). Interestingly, migration speed is not a function of collagen concentration in these tracks (98). PA troughs that confine cells on three sides can also be molded from microfabricated masters. The migration speed of U373-MG human glioma cells through microchannels with a width of 10, 20, or 40 μm and a stiffness ranging from 0.4 to 120 kPa is biphasic in wide (20–40-μm) channels but increases monotonically with stiffness in 10-μm-wide troughs (97).
2.6. Micropipette Aspiration Assay
Several engineering methods can impose well-defined forces on the cell membrane. These methods include magnetic beads (99), stretching devices (100–102), and micropipette aspiration assays (103, 104). Micropipettes are used to generate negative pressures on the cell membrane, resulting in local deformation into the micropipette. The magnitude of the aspiration force is usually controlled by a micromanipulator mounted on a microscopy stage. The applied force commonly ranges from 0.01 to 0.6 nN/μm2, maintaining the integrity of the cell membrane and avoiding cell wounding (103, 104). Micropipette aspiration is used to investigate recruitment or activation of cortical molecules, such as myosin II, cortexillin, and other related cross-linker molecules, following deformation of the cell membrane (103, 104). This assay can also be employed to study the deformation of the nucleus upon applied force (105).
3. CONFINEMENT AND MECHANICAL FORCE INDUCE MOLECULAR CHANGES
The bioengineering tools described above have been used to explore how confinement and mechanical forces influence the biomolecular properties of cells. In particular, physical confinement modulates intracellular molecules responsible for cell morphology, adhesion, contractility, and gene expression.
3.1. Cytoskeletal Architecture Is Reorganized upon Physical Confinement
Physical confinement causes changes in cytoskeletal architecture, typically characterized by the orientation of cytoskeletal components along the axis of cell migration (Figure 2). Confinement inside narrow microchannels tends to cause actin accumulation around the cell cortex, suppression of stress fibers, and orientation of phosphorylated myosin light chain (MLC) along the axis of migration, regardless of substrate stiffness (14, 31, 97, 98). Similarly, on narrow fibronectin printed lines, actin and microtubules are oriented parallel to the printed lines, with stabilized microtubules accumulating in an anterior bundle (65). Vertical confinement in sandwich assays causes a similar reduction of stress fibers, and myosin II tends to concentrate at the cell rear (15). Interestingly, microtubule dynamics appear to be particularly important in confined migration. Inhibition of microtubule polymerization in either narrow microchannels (4, 14, 106) or 1D printed lines (65) significantly reduces migration speed and directionality. In contrast, microtubule polymerization is not required for lobopodia-based migration in 3D CDMs (17).
Figure 2.
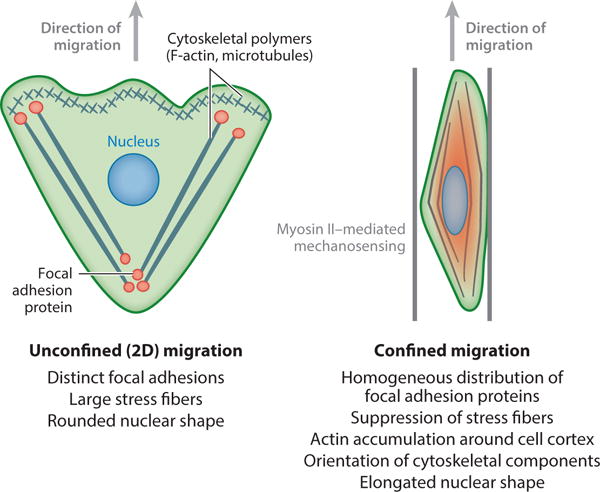
Schematics of cell morphology in unconfined versus confined microenvironments. Cells migrating on planar, two-dimensional (2D) surfaces form large focal adhesions and stress fibers, have rounded nuclei, and often migrate with a broad leading edge containing branching F-actin structures. Conversely, cells in confinement are characterized by a suppression of large focal adhesions and stress fibers. Instead, focal adhesion proteins are often distributed homogeneously around the cell body. Confined cells typically display alignment of cytoskeletal structures and nuclear elongation along the confining axis.
Although intracellular signaling pathways likely play a role in the reorganization of cytoskeletal structure upon confinement, cytoskeletal components can themselves respond to physical forces. For example, experimental and in silico results suggest that extensional but not compressional force promotes Arp2/3-mediated actin filament branching, indicating that confinement drives the direction of cell protrusions (107). Acellular solutions containing actin filaments can also form an actin ring that orients in the plane perpendicular to compressive forces generated by two silicone-coated coverslips spaced approximately 4.5 μm apart (108). This finding suggests that actin spontaneously responds to mechanical force.
3.2. Cellular Adhesion Size Is Reduced upon Physical Confinement
In two dimensions, cells sense and interact with their environment through adhesions with the substrate. Confinement alters the type and morphology of these adhesions. While MDA-MB-231 cells form distinct focal adhesions in 50-μm-wide PDMS microchannels, suppression of focal adhesions and homogeneous expression of phosphorylated focal adhesion kinase (pY-FAK) and pY-paxillin along the cell body occur in 3-μm-wide microchannels (14). Similarly, when fibroblasts migrate on 1D microcontact printed fibronectin lines, α5-integrin, FAK, vinculin, and paxillin are dispersed along the body of the cell instead of being localized at distinct adhesions (65). Vinculin is also homogeneously distributed across the cell body in vertically confined cells (15). Cells moving from narrow to wide microcontact printed lines form larger adhesions upon exiting confining spaces (74). On grooved silicon oxide substrates spaced 70–1,900 nm apart, focal adhesions were also smaller than those seen on 2D surfaces, with focal adhesion width generally increasing as the width of the ridges between the grooves increased (53). Cells migrate efficiently in these environments upon suppression of large focal adhesions, and the reduced dependence of cell speed on ligand density during confinement (65, 98) may be due to the reduced adhesion size observed in confined migration. Taken together, these observations show that confinement causes marked alterations in the cellular cytoskeleton and the pattern of cellular adhesions, suggesting that cells sense and respond to physical confinement.
3.3. Intracellular Signaling via Mechanotransduction Pathways Modulates Cell Response to Confinement
Cells identify and respond to their environment in a process called mechanotransduction (109). Because myosin II is both a sensor and a regulator of cell shape (110, 111), its activity and recruitment are likely to mediate cell response to confinement. The activity of myosin II is regulated by the phosphorylation of MLC (13), which in turn is phosphorylated by MLC kinase (MLCK) and/or Rho kinase (ROCK) (13). ROCK is activated by Rho GTPases, whereas MLCK is regulated by both intracellular calcium and several kinases (13). ROCK and MLCK do not act entirely independently; ROCK can also promote MLC phosphorylation by downregulating myosin phosphatase (112). Myosin II activity and localization are regulated by forces that occur upon cell confinement. For example, compression imposed by vertical confinement induces myosin II activity and localization of actin and myosin II at the rear of the cell cortex, which is critical to mediate the mesenchymal–amoeboid transition (15). Mechanical tension caused by aspiration through a micropipette promotes cortical myosin II recruitment and redistribution in Drosophila embryo cells (103, 113) and in Dictyostelium (114). This recruiting mechanism involves interactive regulation of cortexillin, IQ motif–containing GTPase-activating protein 1 (IQGAP), and myosin II (104, 115, 116). When actin–myosin interactions are interrupted by treatment with blebbistatin, cells exhibit a greatly reduced response to topography (74, 117, 118).
Forces imposed on cells by the external microenvironment may translate to increased myosin activity either through activation of the Rho/ROCK contractility pathway or by calcium influx. Cardiac myocytes, endothelial cells, and NIH-3T3 cells experiencing stretching stress generated by deformable substrates demonstrate increased RhoA/ROCK activation and MLC phosphorylation (102, 119, 120). Magnetic beads bound to integrins can also be used to generate force to stretch the cell membrane, leading to RhoA activation through guanine nucleotide exchange factors (GEFs), GEF-H1, and leukemia-associated Rho GEF (LARG) (99). Conversely, application of 1–5-nN stretching forces on magnetic beads bound to integrins induces rapid calcium influx, in turn modulating cellular function via the calcium–calmodulin pathway (121). Compressive forces regulate calcium influx and oscillation in chondrocytes and myocytes by deforming the cell membrane (122, 123). The calcium–calmodulin complex directly phosphorylates MLCK, leading to myosin II activation in a wide variety of cells (124, 125). Evidence suggests that compensatory mechanisms for myosin II activation exist if either RhoA/ROCK-or MLCK-mediated signaling is inhibited. For example, mechanical compression generated by a weighted piston increases the rate of wound closure in 67NR breast cancer cells (126). Interestingly, cell response to compression is intact upon inhibition of ROCK or MLCK but is lost if contractility is inhibited entirely by treatment with blebbistatin (126), indicating that myosin II can be activated through either of these pathways but must be intact for a mechanosensitive response. However, myosin isoforms may have differential roles in confined migration, as knockdown of myosin IIA but not myosin IIB reduces the migration speed of fibroblasts migrating on 1D printed lines (127) and of Chinese hamster ovary (CHO) cells migrating in narrow microchannels (2).
Although myosin II activity is vital for cell mechanosensitivity, cells in confinement can migrate using a range of mechanisms with varying reliance on cell contractility. On one hand, an amoeboidal migration mode depends strongly on cortical contractility (2, 15). On the other hand, protrusion-based migration modes do not rely on cell contractility in confinement. Some tumor cell lines migrate with the same (or greater) speed as control cells upon inhibition of cell contractility in both stiff and compliant microchannels (14, 97, 98), and force exertion in highly confining PDMS microchannels is not modulated by activation or inhibition of the actomyosin pathway (36). Supporting the notion that contractility and migration speed can be decoupled in confinement is the observation that the phosphorylation level of MLC does not correlate with migration speed in 10-μm-wide, 20-μm-tall microtracks patterned in 3.0 mg/mL collagen gels (98). Evidence suggests that cells can switch between these migration modes, depending on constraints imposed by the microenvironment and modulators of actomyosin contractility. Indeed, highly confined cells in low-adhesion environments can migrate rapidly upon ROCK inhibition in a manner that is dependent on fast actin retrograde flow in a small protrusive region at the cell front, whereas these cells adopt a blebbing mechanism dependent on actomyosin contractility when the contractile pathway is intact (15). Similarly, in 3D CDMs, knockdown of RhoA or inhibition of ROCK switches the adhesion-dependent migration of fibroblasts from lobopodia based to lamellipodia based without affecting migration speed (16). In highly confining microchannels, cells migrate in the absence of actin polymerization by a process dependent on transmembrane water intake at the leading edge and expulsion at the trailing edge (4).
In summary, confinement triggers a series of signaling cascades via mechanotransduction, sequentially regulating cellular behaviors. Our data (W.C. Hung & K. Konstantopoulos, unpublished data) and the fact that both calcium (through MLCK) and RhoA (through ROCK) can independently or cooperatively regulate phosphorylation of MLCK indicate that forces imposed on migratory cells by confinement likely regulate calcium influx, RhoA, and myosin II activities in a complex, intertwined network.
3.4. Gene Expression and Proliferation Are Altered by Mechanical Forces During Confinement
Cell geometry determines the topography of the nucleus, which is critical for the regulation of gene expression through the coupling of histone acetylation and cytoskeleton contractility (128, 129). In confinement, the nucleus assumes a cylindrical shape (Figure 2) through both compressive forces imparted by the microenvironment and stretching forces from the cytoskeleton (2). For example, the nucleus elongates and aligns along microgrooves in fibroblasts plated on 12.5-μm-wide, 2-μm-deep grooves etched in quartz slides (130). On microcontact printed lines, the linker of nucleoskeleton and cytoskeleton (LINC) complex connects the cytoskeleton and the nucleus and, along with actomyosin contractility and a perinuclear actin cap, shapes the nucleus in response to mechanical cues (71, 72). Interestingly, the actin cap maintains nuclear elongation and suppresses nuclear rotation to promote directional cell migration (72).
Cell confinement causes quantifiable changes in gene expression. Cells plated on microgrooves have a number of genes that are upregulated 24 h after plating, including genes involved in Rho GTPase signaling and microtubule organization (130). A large body of evidence suggests that mechanotransduction regulates cell signaling through direct modulation of GTPases, which in turn regulate other cell responses such as survival, division, and proliferation. For example, it is well established that the Rho GTPase family is critical to the regulation of proliferation through its effectors ROCK and mDia, which modulate cell contractility and actin polymerization, respectively (131). Conversely, Rac1 may modulate proliferation through p21-activated protein kinase in the NF-κB pathway (132). Additionally, RhoA activation sequesters the transcription factor YAP/TAZ, a proliferative regulator of the Hippo pathway, in the nucleus to promote proliferation (133). Cells confined to microcontact printed islands designed to cause nuclear elongation also synthesize more collagen I and express higher levels of osteocalcin mRNA than do cells of similar spread area but without nuclear elongation (134). Thus, the physical microenvironment can itself drive changes in gene expression, likely through the action of mechanotransductive signaling pathways.
3.5. Key Differences in Confined Migration Assays
Although 1D microcontact printing assays, microgroove assays, microchannel assays, and vertical confinement assays generate similar cell morphology, there are differences in the migration modes enabled by each type of confinement. First, confinement enables cells to push against their environment and apply forces in the absence of integrin-mediated adhesions (15). Forces in the absence of integrin-mediated adhesion are physiologically important. Blebbing migration of A375M2 melanoma cells occurs in vivo, even when β1-integrin is knocked down (83), and leukocyte migration speed within lymph nodes is not reduced upon knockdown of integrins (135). Although 1D confinement assays recapitulate morphological characteristics of microchannel and 3D migration assays (namely reduced adhesion size and suppression of stress fibers), they depend on adhesion to the substrate. Cells can de-adhere from patterned lines under conditions of high contractility (65). Thus, although these assays provide confinement, they lack important mechanisms of cell motility, such as blebbing, which requires high contractility and low levels of adhesion to the substrate (15).
Second, vertical confinement in sandwich-type assays lacks the full 3D confinement possible in PDMS microchannels or patterned hydrogels. It is currently unknown whether cell migration in microchannels is equivalent to “chimneying” between parallel planes, or whether cells in microchannels actually can exert outward forces on all four walls of a microchannel. Expansion of channel size in patterned hydrogels suggests that force exertion occurs around the entire cell periphery (96), as does evidence that forces exerted by cells confined within PDMS microchannels are oriented toward the microchannel walls (36). It is possible that in very wide but short or very tall but narrow microchannels, only two parallel channel walls are “used” by the cell to move, generating results similar to those observed in vertical confinement assays. However, when confinement increases, cells may adopt alternative migration strategies, such as osmotically driven migration (4). Figure 1 summarizes several of these differences.
In general, the adaptability of migration mechanisms across microenvironments is not fully understood. For example, whereas both lobopodia-based migration and blebbing migration depend on cytoplasmic pressure to move the cell membrane, they differ in that lobopodia-based migration relies on adhesion to the substrate (16). Even among microchannel assays, the use of degradable versus nondegradable polymers to create microtracks for cell migration may yield important differences in cell motility. For example, treatment with blebbistatin does not affect the migration speed of tumor cells in nondegradable PDMS (14) or PA (97) channels, but it decreases migration speed in collagen microtracks (98). It will be important to test migration mechanisms under different types of confinement to discover their generality and elucidate how cells switch between migration modes.
4. TRANSLATIONAL APPLICATIONS OF STUDYING CELL BEHAVIOR IN CONFINEMENT
4.1. Antimetastatic Effects of Modulating the Tumor Microenvironment
The response of tumor cells to the physical microenvironment, which is mediated by the pathways described above, may be exploitable for the treatment of cancer and other diseases. For example, glioblastoma multiforme (GBM) cells preferentially migrate along white matter tracts and blood vessels to invade the brain, making surgical resection difficult (29). Jain et al. (136) exploited GBM cell invasion along topographical cues to draw cells away from a GBM tumor established in a mouse model. An axon guide was filled with an aligned nanofiber membrane and implanted adjacent to a GBM tumor, and U87-MG-eGFP cells preferentially migrated through the guide to significantly reduce the tumor volume in the brain (136). Importantly, the aligned topography of the nanofiber membrane was crucial for this result, as empty axon guides or conduits filled with a smooth (instead of topographically patterned) film did not lead to the same degree of tumor reduction (136). The conduit was paired with a cyclopamine-conjugated collagen hydrogel to selectively cause apoptosis in cells migrating into the conduit and expressing the sonic hedgehog (Shh) pathway, which is overexpressed in most high-grade brain tumors (136).
Along these lines, the mechanosensitive pathways used by cells to sense the physical microenvironment may target cells in regions of dysplasia. A recent study found that patient-derived GBM cells lack the sensitivity to substrate stiffness (e.g., cell rounding and impaired migration on soft substrates) that is observed in the U373-MG GBM cell line, making these cells highly migratory across the stiffness range observed in the brain (137). When these patient-derived cells were transfected with constitutively active RhoA, ROCK, or MLCK, contractility increased and cells regained stiffness sensitivity, presumably due to an increased ability to deform the matrix and sense its stiffness (137). When cells expressing constitutively active RhoA were orthotopically implanted in a mouse xenograft model, survival significantly increased compared with the control case, with a reduction in tumor cell invasion in the brain and tumor size (137). In summary, the use of tumor mechanosensitive pathways and topography-mediated functional behaviors represents an underexplored therapeutic pathway.
4.2. Point-of-Care Microfluidic Devices Using Whole Blood from Patients
The small number of cells used in microfluidic devices makes them especially promising for point-of-care applications. Thus far, these applications, at least in the context of cell migration, have been explored using immune cells. Such devices can handle unprocessed blood at very small volumes (∼2 μL) and have been used to demonstrate decreased neutrophil migration up an N-formyl–methionine–leucine–phenylalanine (fMLP) gradient after burn injuries (138, 139). A similar device can distinguish between burn patients developing sepsis and those not developing sepsis on the basis of the paths taken by neutrophils migrating in a microfluidic device with mazelike microchannels; specifically, the device noted spontaneous migration in the absence of a chemoattractant gradient in patients developing sepsis (140). Importantly, the “score” generated by this device distinguished between patients developing sepsis and those not developing sepsis up to 2 days before clinical diagnosis, suggesting that it can be used as an early aid in determining antibiotic dosing (140).
Such devices may also help discover treatments that rapidly recover proper neutrophil function and migration after burns. For example, the migration efficiency of neutrophils in a microfluidic device correlates with survival in a rat burn model, with neutrophil function in vitro and in vivo largely restored by treatment with resolvin RvD2 following the burn (141). Resolvin treatment also made burned rats less susceptible to microbial insult (141). Similar studies with microfluidic channels have suggested aberrant migration of fibroblasts with a mutation in the TOR1A gene, which underlies early-onset generalized torsion dystonia (142); examined microglial chemotaxis up a soluble amyloid-β gradient in an Alzheimer’s model (143); and elucidated how neutrophil retrotaxis prevents detrimental neutrophil accumulation at sites eliciting an immune response (144).
5. CONCLUSION
The importance of the physical microenvironment in regulating cell signaling and motility has gained increasing recognition in recent years (145). The wide variety of confinement models available to researchers will allow orthogonal control of physical inputs to determine the effect of the microenvironment on cell behavior and fate. Bioengineering models disentangle physical and chemical inputs to determine key regulators and effectors of cell migration and mechanotransduction. Recapitulation of the key aspects of the microenvironment using these models may lead to high-throughput assays that deliver the same conclusions as in vivo studies at a much faster pace. Translation of these findings to the clinic offers the potential of both faster and more accurate tests and treatments for human disease.
In the future, elucidating the precise effects of physical forces on cell behavior will be crucial to delineating the key underlying modes of motility. For example, differential effects of compressive forces and stretching forces on cell proliferation have been reported (146). It will be very important to determine the differential roles of compressive and stretching force in mechanotransduction, as well as the interplay among GTPases, gene regulation, and other mechanosensing mechanisms in confinement. Furthermore, these forces should be defined among various types of confinement assays. Finally, key mechanisms should be tested in various well-characterized assays to determine the features of the microenvironment driving a given result.
Acknowledgments
The writing of this review was supported by grants from the National Institutes of Health (RO1 GM11465) and the National Cancer Institute (RO1 CA183804 and RO1 CA186286).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Hung WC, Chen SH, Paul CD, Stroka KM, Lo YC, et al. Distinct signaling mechanisms regulate migration in unconfined versus confined spaces. J Cell Biol. 2013;202:807–24. doi: 10.1083/jcb.201302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong Z, Balzer EM, Dallas MR, Hung WC, Stebe KJ, Konstantopoulos K. Chemotaxis of cell populations through confined spaces at single-cell resolution. PLOS ONE. 2012;7:e29211. doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stroka KM, Jiang H, Chen SH, Tong Z, Wirtz D, et al. Water permeation drives tumor cell migration in confined microenvironments. Cell. 2014;157:611–23. doi: 10.1016/j.cell.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montell DJ. Border-cell migration: The race is on. Nat Rev Mol Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- 7.Herbert SP, Stainier DYR. Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat Rev Mol Cell Biol. 2011;12:551–64. doi: 10.1038/nrm3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: present and future therapeutic targets. Nat Immunol. 2005;6:1182–90. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 9.Friedl P, Weigelin B. Interstitial leukocyte migration and immune function. Nat Immunol. 2008;9:960–69. doi: 10.1038/ni.f.212. [DOI] [PubMed] [Google Scholar]
- 10.Wrobel MR, Sundararaghavan HG. Directed migration in neural tissue engineering. Tissue Eng B. 2014;20:93–105. doi: 10.1089/ten.TEB.2013.0233. [DOI] [PubMed] [Google Scholar]
- 11.Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat Rev Cancer. 2003;3:921–30. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 12.Ananthakrishnan R, Ehrlicher A. The forces behind cell movement. Int J Biol Sci. 2007;3:303–17. doi: 10.7150/ijbs.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 14.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–56. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu YJ, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, et al. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160:659–72. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J Cell Biol. 2012;197:439–55. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345:1062–65. doi: 10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature. 2014;507:181–89. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 19.Berthier E, Young EW, Beebe DJ. Engineers are from PDMS-land, biologists are from Polystyrenia. Lab Chip. 2012;12:1224–37. doi: 10.1039/c2lc20982a. [DOI] [PubMed] [Google Scholar]
- 20.Duffy DC, McDonald JC, Schueller OJ, Whitesides GM. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Anal Chem. 1998;70:4974–84. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 21.Heuzé ML, Collin O, Terriac E, Lennon-Duménil AM, Piel M. Cell migration in confinement: a micro-channel-based assay. Methods Mol Biol. 2011;769:415–34. doi: 10.1007/978-1-61779-207-6_28. [DOI] [PubMed] [Google Scholar]
- 22.Irimia D, Charras G, Agrawal N, Mitchison T, Toner M. Polar stimulation and constrained cell migration in microfluidic channels. Lab Chip. 2007;7:1783–90. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irimia D, Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol. 2009;1:506–12. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P. Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochem Cell Biol. 2008;130:1147–54. doi: 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 25.Alexander S, Weigelin B, Winkler F, Friedl P. Preclinical intravital microscopy of the tumour–stroma interface: invasion, metastasis, and therapy response. Curr Opin Cell Biol. 2013;25:659–71. doi: 10.1016/j.ceb.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Weigelin B, Bakker G-J, Friedl P. Intravital third harmonic generation microscopy of collective melanoma cell invasion: principles of interface guidance and microvesicle dynamics. IntraVital. 2012;1:32–43. doi: 10.4161/intv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamauchi K, Yang M, Hayashi K, Jiang P, Yamamoto N, et al. Induction of cancer metastasis by cyclophosphamide pretreatment of host mice: an opposite effect of chemotherapy. Cancer Res. 2008;68:516–20. doi: 10.1158/0008-5472.CAN-07-3063. [DOI] [PubMed] [Google Scholar]
- 28.Yamauchi K, Yang M, Jiang P, Yamamoto N, Xu M, et al. Real-time in vivo dual-color imaging of intracapillary cancer cell and nucleus deformation and migration. Cancer Res. 2005;65:4246–52. doi: 10.1158/0008-5472.CAN-05-0069. [DOI] [PubMed] [Google Scholar]
- 29.Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15:455–65. doi: 10.1038/nrn3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 31.Kraning-Rush CM, Carey SP, Lampi MC, Reinhart-King CA. Microfabricated collagen tracks facilitate single cell metastatic invasion in 3D. Integr Biol. 2013;5:606–16. doi: 10.1039/c3ib20196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mak M, Reinhart-King CA, Erickson D. Microfabricated physical spatial gradients for investigating cell migration and invasion dynamics. PLOS ONE. 2011;6:e20825. doi: 10.1371/journal.pone.0020825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mak M, Reinhart-King CA, Erickson D. Elucidating mechanical transition effects of invading cancer cells with a subnucleus-scaled microfluidic serial dimensional modulation device. Lab Chip. 2013;13:340–48. doi: 10.1039/c2lc41117b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pathak A, Kumar S. Transforming potential and matrix stiffness co-regulate confinement sensitivity of tumor cell migration. Integr Biol. 2013;5:1067–75. doi: 10.1039/c3ib40017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prentice-Mott HV, Chang CH, Mahadevan L, Mitchison TJ, Irimia D, Shah JV. Biased migration of confined neutrophil-like cells in asymmetric hydraulic environments. PNAS. 2013;110:21006–11. doi: 10.1073/pnas.1317441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raman PS, Paul CD, Stroka KM, Konstantopoulos K. Probing cell traction forces in confined microenvironments. Lab Chip. 2013;13:4599–607. doi: 10.1039/c3lc50802a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolli CG, Seufferlein T, Kemkemer R, Spatz JP. Impact of tumor cell cytoskeleton organization on invasiveness and migration: a microchannel-based approach. PLOS ONE. 2010;5:e8726. doi: 10.1371/journal.pone.0008726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–87. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weninger W, Biro M, Jain R. Leukocyte migration in the interstitial space of non-lymphoid organs. Nat Rev Immunol. 2014;14:232–46. doi: 10.1038/nri3641. [DOI] [PubMed] [Google Scholar]
- 40.Breckenridge MT, Egelhoff TT, Baskaran H. A microfluidic imaging chamber for the direct observation of chemotactic transmigration. Biomed Microdevices. 2010;12:543–53. doi: 10.1007/s10544-010-9411-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, Chen SH, Hung WC, Paul C, Zhu F, et al. Fluid shear promotes chondrosarcoma cell invasion by activating matrix metalloproteinase 12 via IGF-2 and VEGF signaling pathways. Oncogene. 2015;34:4558–69. doi: 10.1038/onc.2014.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen SH, Hung WC, Wang P, Paul C, Konstantopoulos K. Mesothelin binding to CA125/MUC16 promotes pancreatic cancer cell motility and invasion via MMP-7 activation. Sci Rep. 2013;3:1870. doi: 10.1038/srep01870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Zhang W, Qin L. Mesenchymal-mode migration assay and antimetastatic drug screening with high-throughput microfluidic channel networks. Angew Chem Int Ed Engl. 2014;53:2344–48. doi: 10.1002/anie.201309885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mycielska ME, Djamgoz MB. Cellular mechanisms of direct-current electric field effects: galvanotaxis and metastatic disease. J Cell Sci. 2004;117:1631–39. doi: 10.1242/jcs.01125. [DOI] [PubMed] [Google Scholar]
- 45.Nuccitelli R. A role for endogenous electric fields in wound healing. Curr Top Dev Biol. 2003;58:1–26. doi: 10.1016/s0070-2153(03)58001-2. [DOI] [PubMed] [Google Scholar]
- 46.Huang YJ, Samorajski J, Kreimer R, Searson PC. The influence of electric field and confinement on cell motility. PLOS ONE. 2013;8:e59447. doi: 10.1371/journal.pone.0059447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mak M, Erickson D. Mechanical decision trees for investigating and modulating single-cell cancer invasion dynamics. Lab Chip. 2014;14:964–71. doi: 10.1039/c3lc51173a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scherber C, Aranyosi AJ, Kulemann B, Thayer SP, Toner M, et al. Epithelial cell guidance by self-generated EGF gradients. Integr Biol. 2012;4:259–69. doi: 10.1039/c2ib00106c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ambravaneswaran V, Wong IY, Aranyosi AJ, Toner M, Irimia D. Directional decisions during neutrophil chemotaxis inside bifurcating channels. Integr Biol. 2010;2:639–47. doi: 10.1039/c0ib00011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim DH, Provenzano PP, Smith CL, Levchenko A. Matrix nanotopography as a regulator of cell function. J Cell Biol. 2012;197:351–60. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, Keely PJ. Collagen reorganization at the tumor–stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallego-Perez D, Higuita-Castro N, Denning L, DeJesus J, Dahl K, et al. Microfabricated mimics of in vivo structural cues for the study of guided tumor cell migration. Lab Chip. 2012;12:4424–32. doi: 10.1039/c2lc40726d. [DOI] [PubMed] [Google Scholar]
- 53.Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J Cell Sci. 2003;116:1881–92. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamilton DW, Oates CJ, Hasanzadeh A, Mittler S. Migration of periodontal ligament fibroblasts on nanometric topographical patterns: influence of filopodia and focal adhesions on contact guidance. PLOS ONE. 2010;5:e15129. doi: 10.1371/journal.pone.0015129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D-H, Han K, Gupta K, Kwon KW, Suh K-Y, Levchenko A. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomaterials. 2009;30:5433–44. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oakley C, Brunette DM. The sequence of alignment of microtubules, focal contacts and actin filaments in fibroblasts spreading on smooth and grooved titanium substrata. J Cell Sci. 1993;106:343–54. doi: 10.1242/jcs.106.1.343. [DOI] [PubMed] [Google Scholar]
- 57.Londono C, Loureiro MJ, Slater B, Lucker PB, Soleas J, et al. Nonautonomous contact guidance signaling during collective cell migration. PNAS. 2014;111:1807–12. doi: 10.1073/pnas.1321852111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Worley KE, Shieh D, Wan LQ. Inhibition of cell–cell adhesion impairs directional epithelial migration on micropatterned surfaces. Integr Biol. 2015;7:580–90. doi: 10.1039/c5ib00073d. [DOI] [PubMed] [Google Scholar]
- 59.Vedula SR, Leong MC, Lai TL, Hersen P, Kabla AJ, et al. Emerging modes of collective cell migration induced by geometrical constraints. PNAS. 2012;109:12974–79. doi: 10.1073/pnas.1119313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Théry M, Piel M. Adhesive micropatterns for cells: a microcontact printing protocol. Cold Spring Harb Protoc. 2009;4:1–11. doi: 10.1101/pdb.prot5255. [DOI] [PubMed] [Google Scholar]
- 61.Maiuri P, Terriac E, Paul-Gilloteaux P, Vignaud T, McNally K, et al. The first World Cell Race. Curr Biol. 2012;22:R673–75. doi: 10.1016/j.cub.2012.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alom Ruiz S, Chen CS. Microcontact printing: a tool to pattern. Soft Matter. 2007;3:168–77. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]
- 63.Tan JL, Liu W, Nelson CM, Raghavan S, Chen CS. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 2004;10:865–72. doi: 10.1089/1076327041348365. [DOI] [PubMed] [Google Scholar]
- 64.Desai RA, Rodriguez NM, Chen CS. “Stamp-off” to micropattern sparse, multicomponent features. Methods Cell Biol. 2014;119:3–16. doi: 10.1016/B978-0-12-416742-1.00001-9. [DOI] [PubMed] [Google Scholar]
- 65.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–90. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong S, Guo WH, Wang YL. Fibroblasts probe substrate rigidity with filopodia extensions before occupying an area. PNAS. 2014;111:17176–81. doi: 10.1073/pnas.1412285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma VP, Beaty BT, Patsialou A, Liu H, Clarke M, et al. Reconstitution of in vivo macrophage–tumor cell pairing and streaming motility on one-dimensional micro-patterned substrates. IntraVital. 2012;1:77–85. doi: 10.4161/intv.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang W, Wyckoff JB, Goswami S, Wang Y, Sidani M, et al. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007;67:3505–11. doi: 10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- 69.Leong MC, Vedula SR, Lim CT, Ladoux B. Geometrical constraints and physical crowding direct collective migration of fibroblasts. Commun Integr Biol. 2013;6:e23197. doi: 10.4161/cib.23197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pouthas F, Girard P, Lecaudey V, Ly TB, Gilmour D, et al. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J Cell Sci. 2008;121:2406–14. doi: 10.1242/jcs.026849. [DOI] [PubMed] [Google Scholar]
- 71.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, et al. A perinuclear actin cap regulates nuclear shape. PNAS. 2009;106:19017–22. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim DH, Cho S, Wirtz D. Tight coupling between nucleus and cell migration through the perinuclear actin cap. J Cell Sci. 2014;127:2528–41. doi: 10.1242/jcs.144345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maiuri P, Rupprecht JF, Wieser S, Ruprecht V, Bénichou O, et al. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell. 2015;161:374–86. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 74.Chang SS, Guo WH, Kim Y, Wang YL. Guidance of cell migration by substrate dimension. Biophys J. 2013;104:313–21. doi: 10.1016/j.bpj.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Théry M, Racine V, Piel M, Pepin A, Dimitrov A, et al. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. PNAS. 2006;103:19771–76. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen B, Kumar G, Co CC, Ho CC. Geometric control of cell migration. Sci Rep. 2013;3:2827. doi: 10.1038/srep02827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–95. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 78.Rape A, Guo WH, Wang YL. The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials. 2011;32:2043–51. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tee S-Y, Fu J, Chen CS, Janmey PA. Cell shape and substrate rigidity both regulate cell stiffness. Biophys J. 2011;100:L25–27. doi: 10.1016/j.bpj.2010.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307–16. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fu J, Wang Y-K, Yang MT, Desai RA, Yu X, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–36. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoffecker IT, Guo WH, Wang YL. Assessing the spatial resolution of cellular rigidity sensing using a micropatterned hydrogel–photoresist composite. Lab Chip. 2011;11:3538–44. doi: 10.1039/c1lc20504h. [DOI] [PubMed] [Google Scholar]
- 83.Tozluoglu M, Tournier AL, Jenkins RP, Hooper S, Bates PA, Sahai E. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat Cell Biol. 2013;15:751–62. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- 84.Le Berre M, Zlotek-Zlotkiewicz E, Bonazzi D, Lautenschlaeger F, Piel M. Methods for two-dimensional cell confinement. Methods Cell Biol. 2014;121:213–29. doi: 10.1016/B978-0-12-800281-0.00014-2. [DOI] [PubMed] [Google Scholar]
- 85.Le Berre M, Aubertin J, Piel M. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol. 2012;4:1406–14. doi: 10.1039/c2ib20056b. [DOI] [PubMed] [Google Scholar]
- 86.Rape AD, Kumar S. A composite hydrogel platform for the dissection of tumor cell migration at tissue interfaces. Biomaterials. 2014;35:8846–53. doi: 10.1016/j.biomaterials.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, et al. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–41. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–84. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haeger A, Krause M, Wolf K, Friedl P. Cell jamming: collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim Biophys Acta. 2014;1840:2386–95. doi: 10.1016/j.bbagen.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 90.Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng A. 2011;17:713–24. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.DeForest CA, Anseth KS. Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nat Chem. 2011;3:925–31. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.DeForest CA, Anseth KS. Photoreversible patterning of biomolecules within click-based hydrogels. Angew Chem Int Ed Engl. 2012;51:1816–19. doi: 10.1002/anie.201106463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mosiewicz KA, Kolb L, van der Vlies AJ, Martino MM, Lienemann PS, et al. In situ cell manipulation through enzymatic hydrogel photopatterning. Nat Mater. 2013;12:1072–78. doi: 10.1038/nmat3766. [DOI] [PubMed] [Google Scholar]
- 94.Kloxin AM, Tibbitt MW, Anseth KS. Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms. Nat Protoc. 2010;5:1867–87. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kirschner CM, Anseth KS. In situ control of cell substrate microtopographies using photolabile hydrogels. Small. 2013;9:578–84. doi: 10.1002/smll.201201841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ilina O, Bakker G-J, Vasaturo A, Hofmann RM, Friedl P. Two-photon laser-generated microtracks in 3D collagen lattices: principles of MMP-dependent and -independent collective cancer cell invasion. Phys Biol. 2011;8:015010. doi: 10.1088/1478-3975/8/1/015010. [DOI] [PubMed] [Google Scholar]
- 97.Pathak A, Kumar S. Independent regulation of tumor cell migration by matrix stiffness and confinement. PNAS. 2012;109:10334–39. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carey SP, Rahman A, Kraning-Rush CM, Romero B, Somasegar S, et al. Comparative mechanisms of cancer cell migration through 3D matrix and physiological microtracks. Am J Physiol Cell Physiol. 2015;308:436–47. doi: 10.1152/ajpcell.00225.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guilluy C, Swaminathan V, Garcia-Mata R, O’Brien ET, Superfine R, Burridge K. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat Cell Biol. 2011;13:722–27. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takemoto K, Ishihara S, Mizutani T, Kawabata K, Haga H. Compressive stress induces dephosphorylation of the myosin regulatory light chain via RhoA phosphorylation by the adenylyl cyclase/protein kinase A signaling pathway. PLOS ONE. 2015;10:e0117937. doi: 10.1371/journal.pone.0117937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mizutani T, Kawabata K, Koyama Y, Takahashi M, Haga H. Regulation of cellular contractile force in response to mechanical stretch by diphosphorylation of myosin regulatory light chain via RhoA signaling cascade. Cell Motil Cytoskelet. 2009;66:389–97. doi: 10.1002/cm.20378. [DOI] [PubMed] [Google Scholar]
- 102.Kaunas R, Nguyen P, Usami S, Chien S. Cooperative effects of Rho and mechanical stretch on stress fiber organization. PNAS. 2005;102:15895–900. doi: 10.1073/pnas.0506041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fernandez-Gonzalez R, de Matos Simoes S, Röper JC, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev Cell. 2009;17:736–43. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Srivastava V, Robinson DN. Mechanical stress and network structure drive protein dynamics during cytokinesis. Curr Biol. 2015;25:663–70. doi: 10.1016/j.cub.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stroka KM, Gu Z, Sun SX, Konstantopoulos K. Bioengineering paradigms for cell migration in confined microenvironments. Curr Opin Cell Biol. 2014;30:41–50. doi: 10.1016/j.ceb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Risca VI, Wang EB, Chaudhuri O, Chia JJ, Geissler PL, Fletcher DA. Actin filament curvature biases branching direction. PNAS. 2012;109:2913–18. doi: 10.1073/pnas.1114292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miyazaki M, Chiba M, Eguchi H, Ohki T, Ishiwata S. Cell-sized spherical confinement induces the spontaneous formation of contractile actomyosin rings in vitro. Nat Cell Biol. 2015;17:480–89. doi: 10.1038/ncb3142. [DOI] [PubMed] [Google Scholar]
- 109.Iskratsch T, Wolfenson H, Sheetz MP. Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat Rev Mol Cell Biol. 2014;15:825–33. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fischer RS, Gardel M, Ma X, Adelstein RS, Waterman CM. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr Biol. 2009;19:260–65. doi: 10.1016/j.cub.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Elliott H, Fischer RS, Myers KA, Desai RA, Gao L, et al. Myosin II controls cellular branching morphogenesis and migration in three dimensions by minimizing cell-surface curvature. Nat Cell Biol. 2015;17:137–47. doi: 10.1038/ncb3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J Cell Biol. 2000;150:797–806. doi: 10.1083/jcb.150.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pouille PA, Ahmadi P, Brunet AC, Farge E. Mechanical signals trigger myosin II redistribution and mesoderm invagination in Drosophila embryos. Sci Signal. 2009;2:ra16. doi: 10.1126/scisignal.2000098. [DOI] [PubMed] [Google Scholar]
- 114.Effler JC, Kee YS, Berk JM, Tran MN, Iglesias PA, Robinson DN. Mitosis-specific mechanosensing and contractile-protein redistribution control cell shape. Curr Biol. 2006;16:1962–67. doi: 10.1016/j.cub.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ren Y, Effler JC, Norstrom M, Luo T, Firtel RA, et al. Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin I. Curr Biol. 2009;19:1421–28. doi: 10.1016/j.cub.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kee YS, Ren Y, Dorfman D, Iijima M, Firtel R, et al. A mechanosensory system governs myosin II accumulation in dividing cells. Mol Biol Cell. 2012;23:1510–23. doi: 10.1091/mbc.E11-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saito AC, Matsui TS, Ohishi T, Sato M, Deguchi S. Contact guidance of smooth muscle cells is associated with tension-mediated adhesion maturation. Exp Cell Res. 2014;327:1–11. doi: 10.1016/j.yexcr.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 118.Frey MT, Tsai IY, Russell TP, Hanks SK, Wang YL. Cellular responses to substrate topography: role of myosin II and focal adhesion kinase. Biophys J. 2006;90:3774–82. doi: 10.1529/biophysj.105.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Torsoni AS, Marin TM, Velloso LA, Franchini KG. RhoA/ROCK signaling is critical to FAK activation by cyclic stretch in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2005;289:1488–96. doi: 10.1152/ajpheart.00692.2004. [DOI] [PubMed] [Google Scholar]
- 120.Liu WF, Nelson CM, Tan JL, Chen CS. Cadherins, RhoA, and Rac1 are differentially required for stretch-mediated proliferation in endothelial versus smooth muscle cells. Circ Res. 2007;101:e44–52. doi: 10.1161/CIRCRESAHA.107.158329. [DOI] [PubMed] [Google Scholar]
- 121.Matthews BD, Overby DR, Mannix R, Ingber DE. Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J Cell Sci. 2006;119:508–18. doi: 10.1242/jcs.02760. [DOI] [PubMed] [Google Scholar]
- 122.Haudenschild DR, Chen J, Pang N, Lotz MK, D’Lima DD. Rho kinase–dependent activation of SOX9 in chondrocytes. Arthritis Rheum. 2010;62:191–200. doi: 10.1002/art.25051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ceelen KK, Oomens CWJ, Stekelenburg A, Bader DL, Baaijens FPT. Changes in intracellular calcium during compression of C2C12 myotubes. Exp Mech. 2009;49:25–33. [Google Scholar]
- 124.Sopko NA, Hannan JL, Bivalacqua TJ. Understanding and targeting the Rho kinase pathway in erectile dysfunction. Nat Rev Urol. 2014;11:622–28. doi: 10.1038/nrurol.2014.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Puetz S, Lubomirov LT, Pfitzer G. Regulation of smooth muscle contraction by small GTPases. Physiology. 2009;24:342–56. doi: 10.1152/physiol.00023.2009. [DOI] [PubMed] [Google Scholar]
- 126.Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, et al. Mechanical compression drives cancer cells toward invasive phenotype. PNAS. 2012;109:911–16. doi: 10.1073/pnas.1118910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Doyle AD, Kutys ML, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Micro-environmental control of cell migration—myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. J Cell Sci. 2012;125:2244–56. doi: 10.1242/jcs.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jain N, Iyer KV, Kumar A, Shivashankar GV. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. PNAS. 2013;110:11349–54. doi: 10.1073/pnas.1300801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roca-Cusachs P, Alcaraz J, Sunyer R, Samitier J, Farré R, Navajas D. Micropatterning of single endothelial cell shape reveals a tight coupling between nuclear volume in G1 and proliferation. Biophys J. 2008;94:4984–95. doi: 10.1529/biophysj.107.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CD, Curtis AS. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp Cell Res. 2003;284:274–82. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 131.Mammoto A, Huang S, Moore K, Oh P, Ingber DE. Role of RhoA, mDia, and ROCK in cell shape–dependent control of the Skp2–p27kip1 pathway and the G1/S transition. J Biol Chem. 2004;279:26323–30. doi: 10.1074/jbc.M402725200. [DOI] [PubMed] [Google Scholar]
- 132.Brown JL, Stowers L, Baer M, Trejo J, Coughlin S, Chant J. Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr Biol. 1996;6:598–605. doi: 10.1016/s0960-9822(02)00546-8. [DOI] [PubMed] [Google Scholar]
- 133.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 134.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. PNAS. 2002;99:1972–77. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 136.Jain A, Betancur M, Patel GD, Valmikinathan CM, Mukhatyar VJ, et al. Guiding intracortical brain tumour cells to an extracortical cytotoxic hydrogel using aligned polymeric nanofibres. Nat Mater. 2014;13:308–16. doi: 10.1038/nmat3878. [DOI] [PubMed] [Google Scholar]
- 137.Wong SY, Ulrich TA, Deleyrolle LP, MacKay JL, Lin JM, et al. Constitutive activation of myosin-dependent contractility sensitizes glioma tumor-initiating cells to mechanical inputs and reduces tissue invasion. Cancer Res. 2015;75:1113–22. doi: 10.1158/0008-5472.CAN-13-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hoang AN, Jones CN, Dimisko L, Hamza B, Martel J, et al. Measuring neutrophil speed and directionality during chemotaxis, directly from a droplet of whole blood. Technology. 2013;1:49. doi: 10.1142/S2339547813500040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Butler KL, Ambravaneswaran V, Agrawal N, Bilodeau M, Toner M, et al. Burn injury reduces neutrophil directional migration speed in microfluidic devices. PLOS ONE. 2010;5:e11921. doi: 10.1371/journal.pone.0011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jones CN, Moore M, Dimisko L, Alexander A, Ibrahim A, et al. Spontaneous neutrophil migration patterns during sepsis after major burns. PLOS ONE. 2014;9:e114509. doi: 10.1371/journal.pone.0114509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kurihara T, Jones CN, Yu YM, Fischman AJ, Watada S, et al. Resolvin D2 restores neutrophil directionality and improves survival after burns. FASEB J. 2013;27:2270–81. doi: 10.1096/fj.12-219519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nery FC, da Hora CC, Atai NA, Kim EY, Hettich J, et al. Microfluidic platform to evaluate migration of cells from patients with DYT1 dystonia. J Neurosci Methods. 2014;232:181–88. doi: 10.1016/j.jneumeth.2014.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cho H, Hashimoto T, Wong E, Hori Y, Wood LB, et al. Microfluidic chemotaxis platform for differentiating the roles of soluble and bound amyloid-β on microglial accumulation. Sci Rep. 2013;3:1823. doi: 10.1038/srep01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hamza B, Irimia D. Whole blood human neutrophil trafficking in a microfluidic model of infection and inflammation. Lab Chip. 2015;15:2625–33. doi: 10.1039/c5lc00245a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Agus DB, Alexander JF, Arap W, Ashili S, Aslan JE, et al. A physical sciences network characterization of non-tumorigenic and metastatic cells. Sci Rep. 2013;3:1449. doi: 10.1038/srep01449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lecuit T, Le Goff L. Orchestrating size and shape during morphogenesis. Nature. 2007;450:189–92. doi: 10.1038/nature06304. [DOI] [PubMed] [Google Scholar]


