Figure 1. p63 transcription factors drive cell migration and invasion in late passage human foreskin keratinocytes (HFK) expressing human papilloma virus (HPV)16 E6/E7 genes.
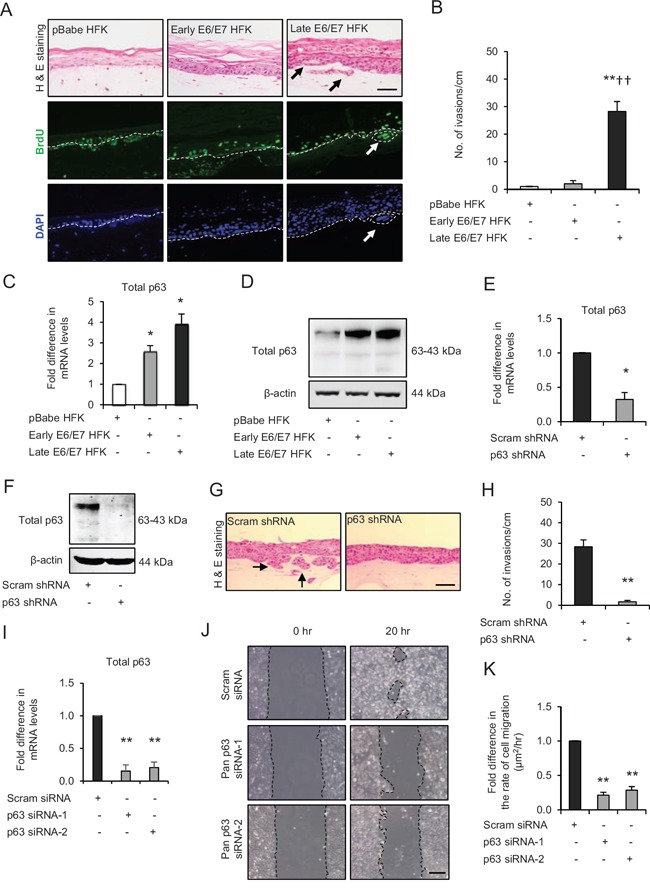
A. H&E staining and immunofluorescence detection of BrdU uptake and nuclei (DAPI) on 3D-organotypic rafts established from normal HFK expressing pBabe (control), early passage and late passage HFK expressing E6/E7 genes cells seeded on retinoblastoma-depleted human foreskin fibroblast embedded collagen-I plugs. Invasive incidents are indicated by arrows. B. Quantification of number of invasive incidents across the rafts per cm. C. The relative mRNA and D. protein expression of total p63 and loading control β-actin in aforementioned cells. E. Relative mRNA and F. protein levels of total p63 in late passage E6/E7-HFK after stable p63 (p63 shRNA) knockdown compared to its controls (scram shRNA). G. H&E staining of rafts established from late passage HFK expressing control and stable p63 knockdown. H. Quantification of number of invasive incidents per cm across the rafts. I. Relative mRNA levels of total p63 after transient knockdown by control and two different p63 siRNA molecules in late passage E6/E7-HFK. J. Representative phase contrast images of scratch wound assay, showing cell migration after 20 hr in these cells seeded on collagen-I coated plates. K. Quantification of migration represented as μm2/hr. Scale bars represent 100 μm. N = 3 independent experiments, mean ± SEM, **p < 0.01 compared to the respective controls, *p < 0.05 compared to the control, ††p < 0.01 compared to early passage E6/E7-HFK.
