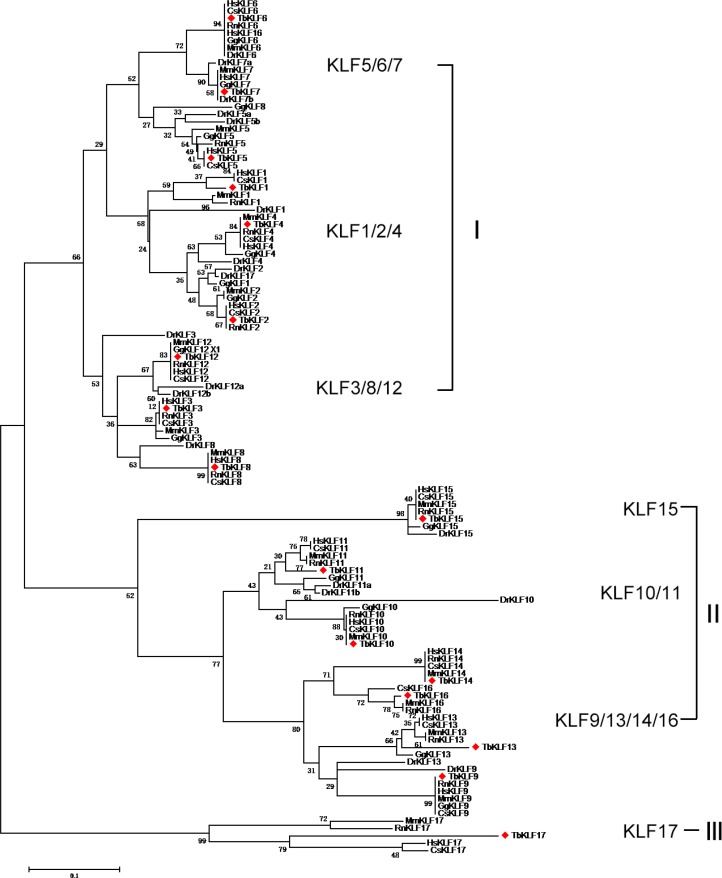
Figure 1. Maximum Likelihood Phylogenetic tree of KLFs from human, monkey, rat, mouse, tree shrew, chicken and zebrafish sequences.
The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The analysis involved 116 amino acid sequences. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA5. The species acronyms are Hs, Homo sapiens (human); Dr, Danio rerio (zebrafish); Mm, Mus musculus (mouse); Gg, Gallus gallus (chicken); Tb, Tupaia belangeri chinensis (tree shrew); Chlorocebus sabaeus (green monkey); and Rattus norvegicus (rat).