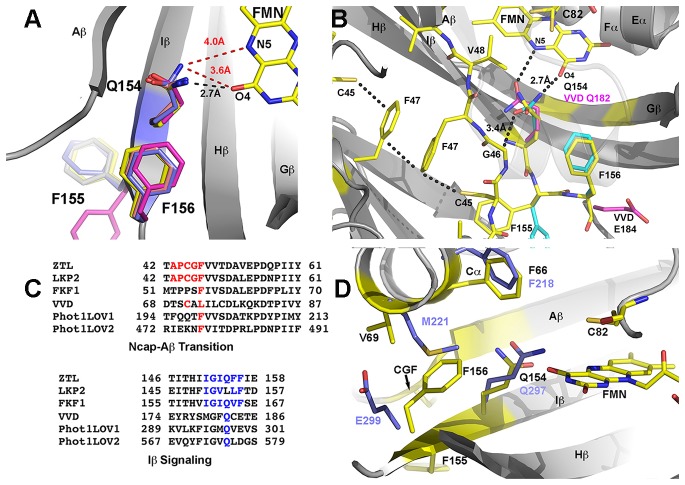
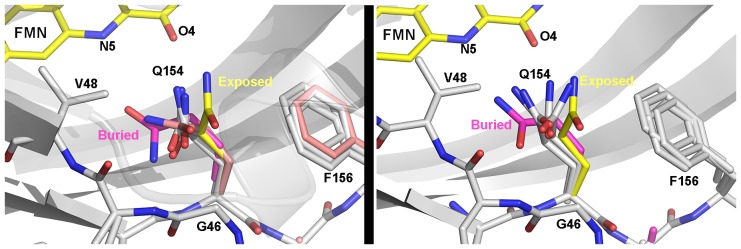
Figure 3. Q154 links Ncap, Ccap and helical elements.
(A) Q154 exists in multiple conformations in WT ZTL structures. They differ in interactions with the active site flavin. An exposed conformation forms strong H-bonds to the O4 position (black dotted line). A buried conformation forms weaker interactions (3.6 Å) with O4 that leads to closer interactions at N5 (4.0 Å; red dotted line). The altered conformation is coupled to movement of F156 into the active site and multiple conformations of F155, forming a QFF motif. The altered conformations define ZTL signaling as distinctly different than existing LOV structures. (B) The unusual orientations of Q154 differ from other LOV proteins that typically show strong interactions near N5 (VVD; magenta). The heterogeneous conformations of Q154 directly abut G46 in a CGF motif allowing formation of the sulfur-π and π-π interactions involving C45 and F47. F156 then adopts a buried conformation in contrast to the equivalent residue in VVD (E184) (C) Sequence alignments of LOV proteins depict conserved elements within the CGF motif (red) and QFF motif (blue) in Arabidopsis thaliana ZTL, LKP2, FKF1, phototropin 1 LOV1 and LOV2. Sequence conservation indicates divergent signaling mechanisms within the ZTL/FKF1 family compared to existing LOV allostery models. (D) Comparisons of ZTL (yellow, black lettering) and Arabidopsis thaliana phototropin 1 LOV1 (PDB: 2Z6C; blue). The altered conformation of Q154 draws F156 into the active site. The buried conformation of F156, leads to movement of Cα (F66, V69). (Figure 3—figure supplement 1)..