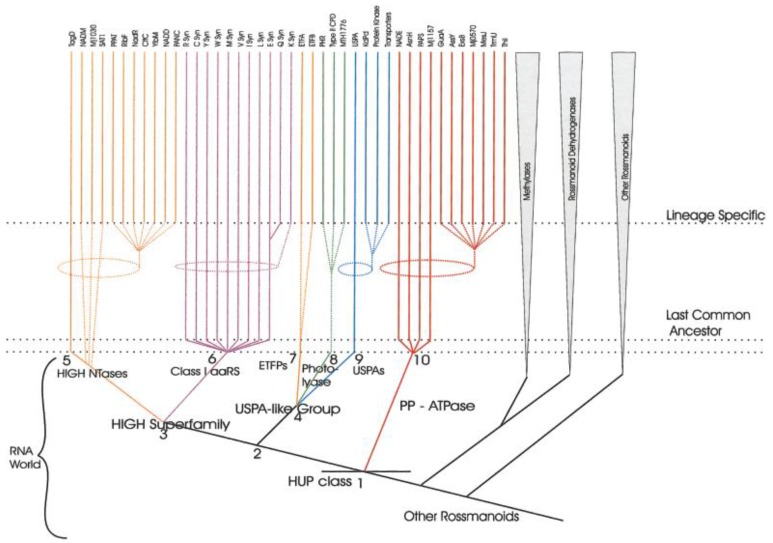
Figure 8.
Evolutionary scenario (cladogram) for the HUP domain class (HIGH-signature proteins, UspA, and PP-ATPase). The specific characters associated with each of the nodes as derived through the cladistic analysis are as follows. Node 1 (the HUP domain): presence of a core 5-strand sheet in the 3–2–1–4–5 order; Node 2: configuration of the region between the last helix and strand of the HUP domain, strand 4 and 5 hydrogen-bonded through most of their lengths; Node 3: HIGH motif in the loop between strand 1 and helix 1, bihelical extension C-terminal to the core HUP domain, with a secondary role in nucleotide-binding; Node 4: loop between strand 3 and helix 4 tends to face outward, helix 4 tends to be placed behind the central sheet, no strongly conserved motif between strand-1 and helix-1, possible loss of ability to hydrolyze α–β bond in ATP; Node 5: nucleotidyl transferase activity, two small residues in the KMSKS loop; Node 6: AARS activity, classical KMSKS motif; Node 7: insertion of a β-hairpin between helix 4 and strand 5; Node 8: fusion with large, α-helical C-terminal domain; Node 9: distinct loop between strand 4 and helix 4; Node 10: SXGXDS motif between strand-1 and helix-1, conserved helix N-terminal to strand 1. The conserved families and their probable temporal points of origin are shown for each of the six major lineages. Dotted ellipses and lines leading to a particular lineage indicate uncertainty regarding its emergence. Reproduced with permission from ref. [73].