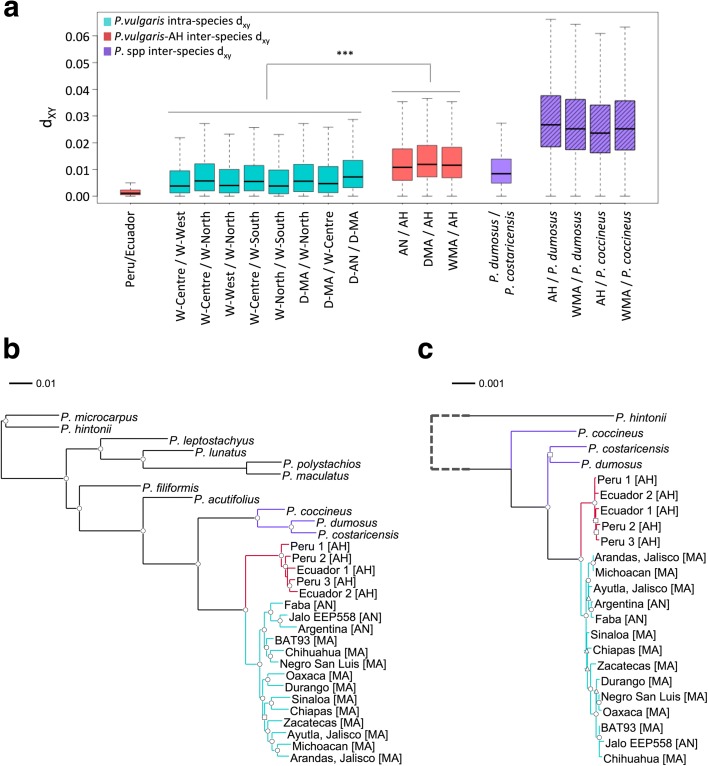
Fig. 1.
Species definition within the Vulgaris group according to their phylogenomic profile. a Absolute genetic divergence between Phaseolus subpopulations, showing inter-species and intra-species divergence comparisons. The difference of dXY values (Kruskal-Wallis p value = 0.014) calculated within P. vulgaris subpopulations and between P. vulgaris and the AH subpopulation, is highlighted with (***). b ML tree with non-parametric SH branch support based on 460,000 single nucleotide polymorphisms randomly chosen across the genome. c ML tree with non-parametric SH branch support based on 55 Kb of the chloroplast genome. The long branch length separating P. hintonii from the Vulgaris species is graphically represented with a dotted line. Branch support: SH-aLRT = [0.75;0.85], triangles; SH-aLRT = [0.85;0.95], squares; SH-aLRT > 0.95, circles. In both tree topologies and the box plot, P. vulgaris accessions are highlighted in cyan, P. pseudovulgaris in red and Phaseolus species from the Vulgaris groups in purple