Abstract
Sine oculis homeobox homolog 1 (Six1) is crucial in normal organ development. Recently, Six1 is reported to display aberrant expression in various cancers and plays important roles in cancer development. However, the regulatory mechanism of Six1 in gastric cancer is largely unknown. In the current study, we found that Six1 was increased in gastric cancer tissues, and its upregulation significantly associated with lymph node metastasis (p=0.042) and poor differentiation (p=0.039). Next, we took advantage of public available microarray data to assess Six1 prognostic value with online K-M Plotter software in gastric cancer, which demonstrated that patients with higher Six1 expression had shorter survival time (p=0.02). To explore the underlying mechanism of Six1, we silenced its upregulation in gastric cells to detect cellular functions. Our results indicated that knock-down Six1 could decrease colony formation number and rendered cells sensitive to 5- Fluorouracil drug treatment. The flow cytometry analyses showed that Six1 silence could promote apoptosis but had little effect on cell cycle transition. Along this clue, we tested mitochondrial membrane potential with JC-1 assay, which suggested that Six1 inhibition could trigger mitochondrial apoptosis. Our subsequent results revealed that Six1 knock-down could reduce the level of anti-apoptotic protein Bcl-2, and caspase-7 but not caspase-3 was involved to execute the mitochondrial apoptosis pathway. Taken together, we find Six1 has oncogenic role in gastric cancer development, and silenced Six1 expression can promote mitochondrial apoptosis by repressing Bcl-2 and activating executor caspase-7. These findings suggest that Six1 may become a valuable prognostic and therapeutic target in gastric cancer.
Keywords: Six1, Gastric cancer, Mitochondrial apoptosis, Caspase-7, Bcl-2
Introduction
As one of leading causes of cancer-related death, most gastric cancer cases are frequently diagnosed at the advanced stage. Surgical resection and chemoradiotherapy are routine approaches, but the therapeutic effects are still limited. So it is urgently needed to investigate the mechanisms and identify more candidate targets for gastric cancer therapy.
Sine oculis homeobox homolog 1 (Six1) is a homeodomain-containing transcription factor which belongs to the Six family. Six1 expresses in various tissues highly through the whole embryogenesis process. In the myogenesis of embryonic skeletal, Six1 expresses before MRFs (myogenic determination factors) and can directly control MRFs expression1. Overexpressed Six1 can expand the preplacodal ectoderm during the growth of the neural crest and epidermis2. The function of Eya1- Six1 complex is essential for consummating the function of auditory organ, because Six1 mutation is able to affect the interaction of both sides, subsequently cause branchio-oto-renal syndrome3. Through binding and increasing mTOR protein directly, the combination of Six1 and Eya2 can regulate physiological myocardial hypertrophy 4.
Many researches have demonstrated that Six1 plays important roles in tumorigenesis. Six1 involves in the genesis, progression and metastasis of multiple cancers, such as cervical cancer5, hepatocellular carcinoma6 and background Wilms tumors7. In cervical cancer, Six1 can enhance TGF-β-induced activation of SMAD2/3, and coordinate with the SMAD pathway to elevate VEGF-C expression, subsequently promote tumor lymphangiogenesis and lymph node metastasis 8. Six1 is also reported to be upregulated in gastric cancer and closely associated with poor prognosis9. This result implicate that Six1 may also exert crucial functions in gastric cancer development. However, so far as we know, there is little study to report Six1 functions, and its underlying mechanisms are largely unknown in gastric cancer.
In our study, we established that Six1 was overexpressed in gastric cancer tissues, which significantly correlated with poor prognosis. Online K-M Plotter analysis indicated that high Six1 cases had shorter overall survival outcome. Moreover, we firstly found that Six1 could modulate mitochondrial apoptosis via Bcl-2 and caspase-7 in gastric cancer cells. Our findings shed new light on Six1 regulatory mechanism and is helpful to develop novel prognostic and therapeutic target in the Six1 pathway.
Materials and Methods
Clinical tissue specimens from gastric cancer and gastritis patients
A total of 40 gastric cancer and 15 gastritis patients were enrolled in this study. 40 patients received curative resection for gastric cancer at general department of Huashan Hospital of Fudan University between November 2012 to March 2014. 15 gastritis specimens were obtained via endoscopic examination. Pathological diagnoses were confirmed by pathologists. After informed consent of the tissue donors, specimens were collected after stomach resection or endoscopic biopsy within 30min and stored immediately in liquid nitrogen until RNA or protein extraction. The collection of clinical specimens was approved by the Ethics Committee of Fudan University.
Online Kaplan Meier plotter (K-M plotter) analysis
The K-M plotter is an online database(www.kmplot.com), which contain gene expression data and survival information downloaded from GEO (Affymetrix microarrays only), EGA and TCGA 10. K-M plotter software can provide online survival analysis of breast, ovarian, lung and gastric cancers with these available transcriptome data. In this website, entire gastric cancer database includes 1,065 gastric cancer samples, and the gene transcriptional levels were measured with the Affymetrix Human Genome U133 Array. In the gastric cancer database, the mean follow-up time was 33 months 11. The Affymetrix probe ID:228347 at Six1 were used to assess the prognostic value of Six1 in gastric cancer overall survival.
Cell culture
Human gastric cancer cell line SGC-7901 was purchased from the Chinese Academy of Sciences (Shanghai, China). Cell was cultured and maintained in RPMI-1640 medium (Invitrogen, 22400089) supplemented with 10% fetal bovine serum (Invitrogen, 10099141) and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin) in a humid atmosphere with 5% CO2 at 37 °C.
Transfection
The small interfering RNA (siRNAs) transfection was conducted when cells grown up to 50% confluency with lipofectamine 2000 (Invitrogen, California, USA) according to manufacture recommendations. After transfection, cells were incubated for another 48 h before being harvested for RNA extraction or 72 h for protein extraction. The siRNA duplexes for Six1 are as follows:
Six1-Si-1: 5'-GCAACUUCCGUGAGCUCUATT-3';
Six1-Si-2: 5'-GUCAGCAACUGGUUUAAGATT-3';
Negative control (NC): 5'-UUCUCCGAACGUGUCACGUTT-3'.
Cell cycle and apoptosis analysis
After washing with PBS, fixed cells were stained with BD Pharmingen™ PI/RNase staining buffer (BD Pharmingen, 550825) for 30min in the dark. PE Annexin V Apoptosis Detection Kit I (BD Pharmingen,559763) was used in apoptosis analysis. After washing with PBS, cells were resuspended in 1X binding buffer at a concentration of 1x106 cells/ml. Then, 5 μl of PE Annexin V and 5 μl 7-AAD were added to 100 μl cell suspension. After 15min incubation in the dark, another 400 μl 1X binding buffer was added. Both in regards to the cell cycle and to apoptosis analysis, the stained cells were analyzed using BD FACS CantoTM Flow Cytometer (BD Biosciences, San Jose, CA, USA).
RNA extraction, reverse transcription and quantitative real time-polymerase chain reaction (qRT-PCR)
The total RNA was extracted from cultured cell line using TRIzol reagent. Reverse transcription was conducted with the PrimeScript™ RT reagent kit (TaKaRa, RR037A). SYBR Premix EX Taq™ II kit (TaKaRa, RR820A) with an ABI PRISM 7500 HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) was used to amplify cDNA template in qRT-PCR. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was adopted as an endogenous control. The primers are as follows:
Six1-RT-F: CGCTCATGTCCAGCTCAGAA;
Six1-RT-R: TAGTTTGAGCTCCTGGCGTG;
GAPDH-RT-F: TCGACAGTCAGCCGCATCTTCTTT;
GAPDH-RT-R: ACCAAATCCGTTGACTCCGACCTT.
Western blot assay
The protein content was measured with BCA Protein Assay Kit (Beyotime, P0010). Protein mixed with 5X loading buffer was boiled at 100 °C for 10 min. After electrophoretically separated on SDS-PAGE (Sangon Biotech, SD6013), the protein was transferred onto nitrocellulose membrane. Then, blocking the membrane in 5% non-fat milk for 1 h at room temperature and then incubating it in monoclonal antibody at 4°C overnight. After three times wash, the membrane was incubated with a fluorescence marked secondary antibody for 1 h at room temperature in the dark. The bands of Western blot were scanned using the Odyssey system (LI-COR, USA) and the bands density was quantified with software Quantity One (Version 4.4.1).The antibodies used in Western blot were listed as follows: anti-Six1 ( #12891,CST, USA), and anti-β-actin (#12620, CST, USA), anti-caspase-7 (#12827, CST, USA), anti-caspase-3 (#9665, CST, USA), anti-cleaved-PARP (#5625, CST, USA), anti-Bcl-2 (#2870, CST, USA), anti-Bax (#5023, CST, USA) and p53 (#sc-126, Santa, USA).
Immunohistochemistry (IHC) Assay
Routine formalin-fixed, paraffin-embedded tissue were cut and mounted on slide. Tissue sections were deparaffinized using xylene and dehydrated with graded ethanol. Citric acid buffer was used for antigen retrieval at 120°C for 20 min. Then, after blocking nonspecific binding sites by exposure in 10% bovine serum albumin, the sections was incubated in anti-Six1(1:200 dilution) antibody at 4°C overnight, followed by 1 h incubation with horseradish peroxidase-conjugated anti-rabbit IgG as second antibody at room temperature, and then visualized using DAB. Counterstaining was conducted with hematoxylin.
Immunofluorescence (IFC) staining Assay
SGC-7901 cells were cultured on coverslips and transfected with Six1 siRNA for 48 h. Then, the cells was fixed in 4% paraformaldehyde for 30 min and permeabilized by 0.25% Triton X-100 for 15 min. After 1 h blocking with 10% normal goat serum, cells were incubated with Six1 antibody at 4°C overnight. Sequentially, the coverslips were treated with Alexa Fluor® 488 goat anti-rabbit IgG secondary antibody for 1 h and followed by incubation with DAPI for 30 min in dark box at room temperature. All images were captured using Nikon fluorescent microscope.
Colony formation assay
Cancer cells SGC-7901 were seeded into 12-well plates at a density of 1500 cells/well in 1 mL RPMI-1640 after transfection for 24 h. Then, incubate the cells at 37°C in a humidified atmosphere with 5% CO2 for 7 days or until colonies appeared. The cells were fixed in 4% methanol and stained with 800µl of Crystal violet to count the colonies.
Cell proliferation assay
SGC-7901 cells were divided into 96-well plates after knocking down Six1 for 24 h, 4000 cells were plated in each 96-well with 100 µl RPMI-1640 and attached to each other overnight. Then, various concentrations of 5- Fluorouracil (5-Fu) was added to treat cells for 48 h. Cell viability was detected by CCK-8 assays, adding 10ul CCK-8 into the media and mixed by gentle agitation. The plate was subsequently incubated for 2 h then the measurement was taken for absorbance under a reference of 450nm.
Mitochondrial membrane potential
The mitochondrial membrane potential (Δψm) was detected by using JC-1 staining method. Briefly, 72h after transfection of Six1 siRNA, cells were harvested, washed twice by PBS and incubated with 5 μM JC-1 dye for 20 min at 37 °C and 5% CO2 in dark. Then, cells were washed twice, resuspended by 1x staining buffer and been observed by fluorescence microscopy or analyzed using a FACS flow cytometer (BD Biosciences, San Jose, CA). Data was analyzed using FlowJo software (Tree Star Inc, Ashland, OR).
Statistical analysis
Statistical analyses were performed with the SPSS statistical package (SPSS, Inc., Chicago, IL, USA). The significance between Six1 expression level and clinical pathological parameters were analyzed using the Mann-Whitney U test. P<0.05 were considered to be statistically significant.
Results
Six1 expression is increased in gastric cancer samples and has clinical significance
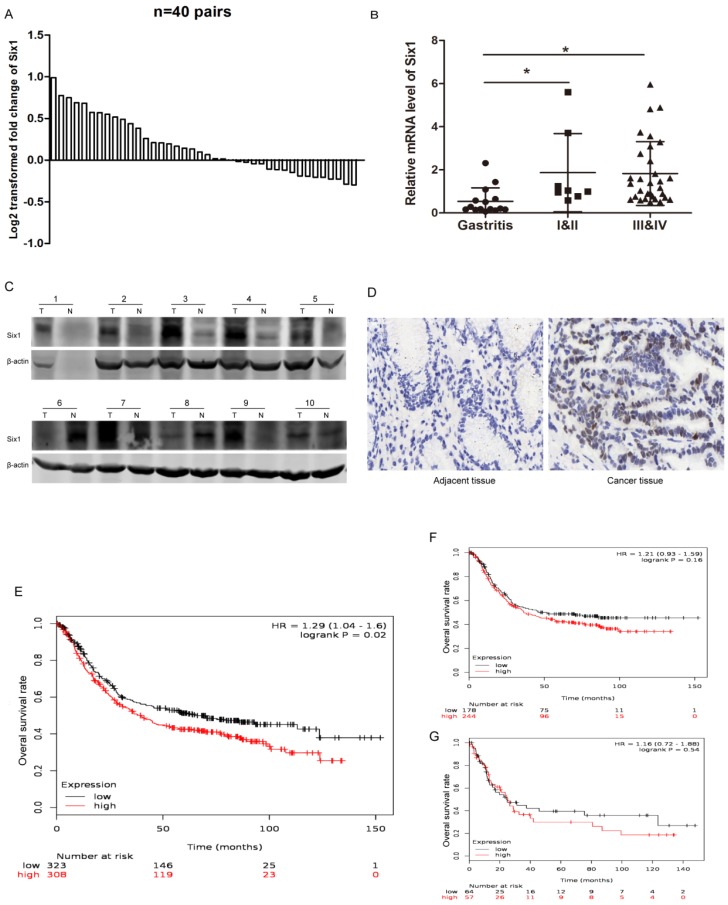
Six1 has been reported to be upregulated in multiple cancers, but it has not been systematically studied in gastric cancer. To establish the expression pattern of Six1 in gastric cancer, we collected 40 paired gastric cancer specimens and detected its transcriptional levels. The qRT-PCR result showed that 23 paired (57.5%) gastric cancer tissues had higher Six1 mRNA levels than its corresponding counterparts (Fig.1A). Statistical analysis demonstrated that Six1 overexpression were dramatically associated with lymph node metastasis (p=0.042) and poor differentiation (p=0.039) (Table 1). Additionally, we analyzed Six1 mRNA levels in gastritis and different TNM stage gastric cancer patients. As shown in Fig. 1B, compared with gastritis cases, Six1 transcriptional levels remarkably increased in gastric cancer patients, despite of no significant differentiation between I&II and III&IV groups. To further confirm Six1 expression, Western blot and IHC assay were carried out in our available paired gastric cancer specimens. Six1 protein exhibited abundant expression in seven out of ten gastric cancer samples (Fig. 1C), and positive Six1 staining were observed in the nuclear of gastric cancer tissues whereas its adjacent counterpart presented little Six1 protein signal (Fig. 1D).
Figure 1.
Six1 expression is increased in gastric cancer samples and has clinical significance. (A) qRT-PCR results showed Six1 mRNA levels in 40 paired gastric cancer tissue specimens. GAPDH was the internal control. The relative fold change of the Six1 mRNA level was performed log2-transformed. (B) Relative mRNA levels of Six1 in gastritis (n=15), TNM I&II (n=8) and TNM III&IV (n=32) specimens respectively. (C) Western blot assay was conducted to detect Six1 protein expression in 10 paired gastric cancer tissues. T was the tumor samples and N was its adjacent normal counterpart. (D) IHC staining evaluated Six1 expresson and distribution in paired gastric cancer samples. (E) Online K-M plotter was used to conduct survival analysis of Six1 in gastric cancer with 631 public available microarray data. (F) Survival analysis of Six1 in 422 samples with lymphatic metastasis. (G) Survival analysis of Six1 in 121 transcriptome date with poor differentiation. The low and high Six1 subgroups were divided according to the median value of Six1 levels. The default parameters were used for analysis. Logrank p < 0.05 was considered to be statistically.
Table 1.
The association of Six1 levels with clinical pathological features in gastric cancer.
| Characteristics | Number | Low | High | p-value |
|---|---|---|---|---|
| Age (years) | ||||
| <=60 | 23 | 9 | 14 | 0.232 |
| >60 | 17 | 9 | 8 | |
| Gender | ||||
| Male | 23 | 14 | 9 | 0.002* |
| Female | 17 | 3 | 14 | |
| Tumor size (cm) | ||||
| <=3cm | 14 | 6 | 8 | 0.967 |
| >3cm | 26 | 11 | 15 | |
| Lymphatic Metastasis | ||||
| Yes | 28 | 8 | 20 | 0.042* |
| No | 12 | 9 | 3 | |
| Differentiation | ||||
| Well&Moderate | 8 | 6 | 2 | 0.039* |
| Poor | 32 | 11 | 21 | |
| Distal metastasis | ||||
| M0 | 38 | 16 | 22 | 0.926 |
| M1 | 2 | 1 | 1 | |
| TNM | ||||
| I&II | 8 | 4 | 4 | 0.908 |
| III&IV | 32 | 13 | 19 |
The Mann-Whitney U test was used to analyze the association and p < 0.05 was statistically significant.
Furthermore, we employed online survival analysis software K-M plotter to assess Six1 prognostic value in gastric cancer. Among the entire 1,065 recruited gastric cancer specimens in the database, Six1 expression was probed in 631 samples. These 631 recruited samples were divided into low and high subgroups according to the median value of Six1 expression levels. As shown in Fig.1E, high Six1 cohort had shorter overall survival time (p=0.02). Among the lymphatic metastasis and poor differentiation subgroups, patients with Six1 increment were prone to have poor survival outcome, despite lack of statistical significance (Fig.1F, G). These results implied that Six1 may play important roles in gastric cancer development and its mechanisms are needed to be explored.
Six1 knockdown can inhibit clone formation and sensitize cells to 5-Fu treatment
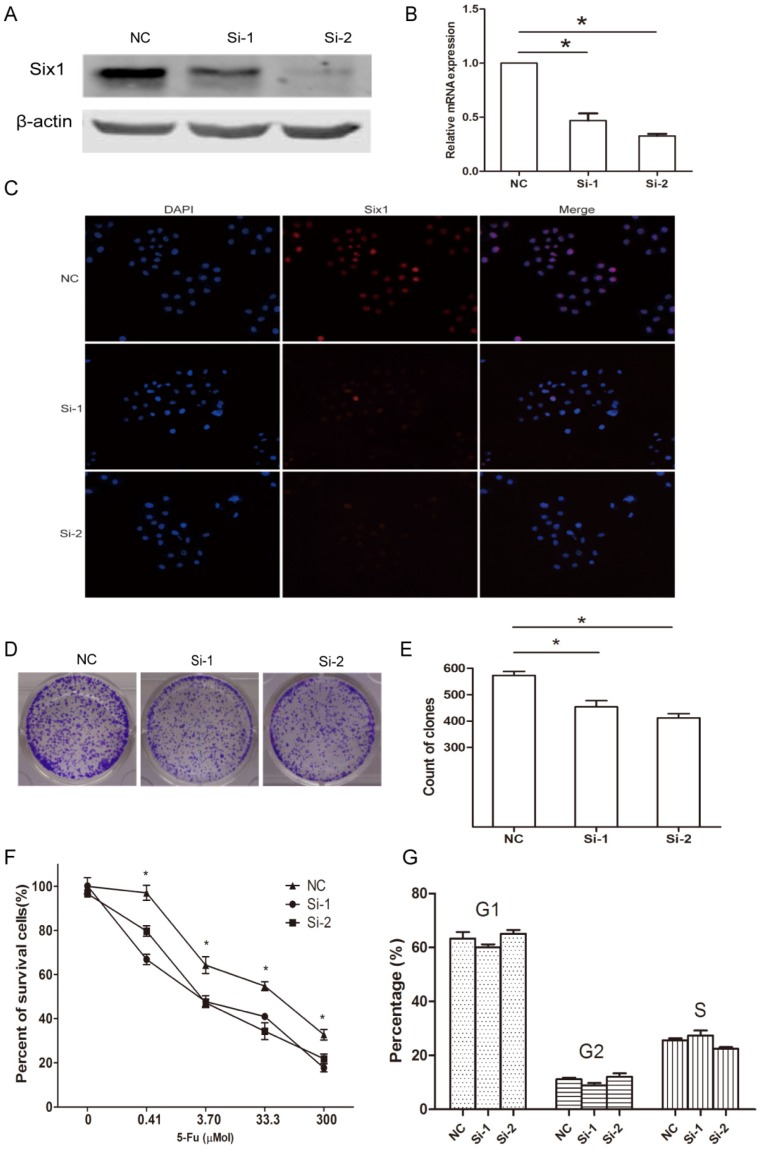
Next, we synthesized two siRNA oligoes to silence Six1 expression in SGC-7901 cells, and assessed its influence on cellular behavior. Western blot assay and qRT-PCR manifested that Six1 was successfully knocked down (Fig. 2A, B). The cellular IFC staining revealed that Six1 mainly localized in the nuclear (Fig. 2C), which was consistent with our IHC result in Fig. 1D. Meanwhile, Six1 fluorescence intensity was weakened apparently with its knock-down (Fig. 2C). Colony-formation assay showed that reduced Six1 expression could decrease the number of survival clone (Fig. 2D, E). When SGC-7901 cells were treated with increasing concentration of chemotherapeutic drugs 5-Fu, cell viability was reduced obviously in a dose-dependent manner, but Six1 silence could have cells more sensitive to 5-Fu (Fig. 2F), which implicated that Six1 upregulation may be partly responsible for 5-Fu resistance in gastric cancer. Then, flow cytometry analysis was carried out to test if Six1 affected cell proliferation via altering cell cycle progression. We found that Six1 knock down had little influence on cell cycle transition (Fig. 2G).
Figure 2.
Six1 knockdown can inhibit clone formation and sensitize cells to 5-Fu treatment. (A) Western blot assay demonstrated that Six1 protein level was decreased with siRNA treatment. β-actin was used as internal control. (B) Six1 transcriptional level was knocked down successfully after siRNA transfection with qRT-PCR detection. (C) IFC staining confirmed Six1 protein was decreased in nuclear after siRNA transfection. (D) The representative picture of clone formation with Six1 silence. (E) The histogram indicated the average number of survival clones in NC and Six1 knock-down groups. (F) Cell viability was measured by CCK-8 assays. The percentage of survival cells reduced more significantly in Six1 knock-down groups when various dose 5-Fu drugs were added. (G) Cell cycle transition was analyzed by flow cytometry. The percentage of each cell cycle phase was not affected by Six1 reduction. Data were shown as mean ± SD (n=3), and *, p < 0.05.
Taken together, these findings suggested that Six1 can affect cell proliferation and 5-Fu resistance but these effects did not result from cell cycle alteration.
Six1 silence can trigger apoptosis especially mitochondrial pathway apoptosis
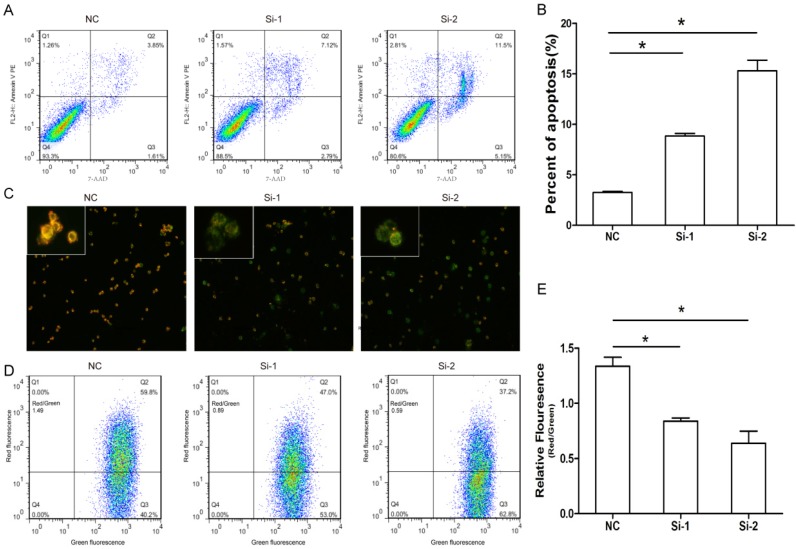
Given Six1 silence had slight effect on cell cycle, we next evaluated whether Six1 could impact on apoptosis. The results showed there were obviously more apoptotic cells in two knockdown groups compared to that in negative control (Fig. 3A, B). These results suggested that Six1 could affect cell proliferation by regulating apoptosis instead of cell cycle progression.
Figure 3.
Six1 silence can trigger apoptosis especially mitochondrial pathway apoptosis. (A) Six1 knock-down promoted apoptosis in SGC-7901 cells. Apoptotic cells were stained with Annexin V and 7-AAD. (B) Histogram showed the percent of apoptotic cells in NC and Six1 knock-down groups. (C) Six1 silence decreased the mitochondrial membrane potential (Δψm) and triggered mitochondrial apoptosis detected by JC-1 assay. JC-1 displays red fluorescence in cells with high Δψm, whereas JC-1 exhibits green fluorescence in cells with low Δψm. (D) Flow cytometry showed the distribution ratio of JC-1 red and JC-1 green cell population. (E) Histogram calculated the relative ratio of red against green fluorescence. *, p < 0.05.
Previous study indicated that cancer development is frequently regulated by mitochondrial apoptosis pathway. So we assessed if Six1 was able to control mitochondrial apoptosis by detecting the variation of Δψm. As we know, the low Δψm can increase permeability of the mitochondrial membrane and promote apoptotic effectors release to initiate apoptosis. The cytofluorimetric dye JC-1 can enter the mitochondria, and form complexes with intense red fluorescence in cell with high Δψm, but in low Δψm cells, JC-1 exhibits green fluorescence. As shown in Fig. 3C, when Six-1 was silenced, SGC-7901 cells had lower Δψm, as the Six-1 knock down exhibited more green fluorescence than control group. Quantitative analysis by flow cytometry verified that the percentage of cells with lower of Δψm (higher ratio of green to red fluorescence) significantly increased after transfecting Six1 siRNA (Fig. 3D, E). These results indicate that knocking down Six1 can trigger mitochondrial apoptosis by decreasing Δψm.
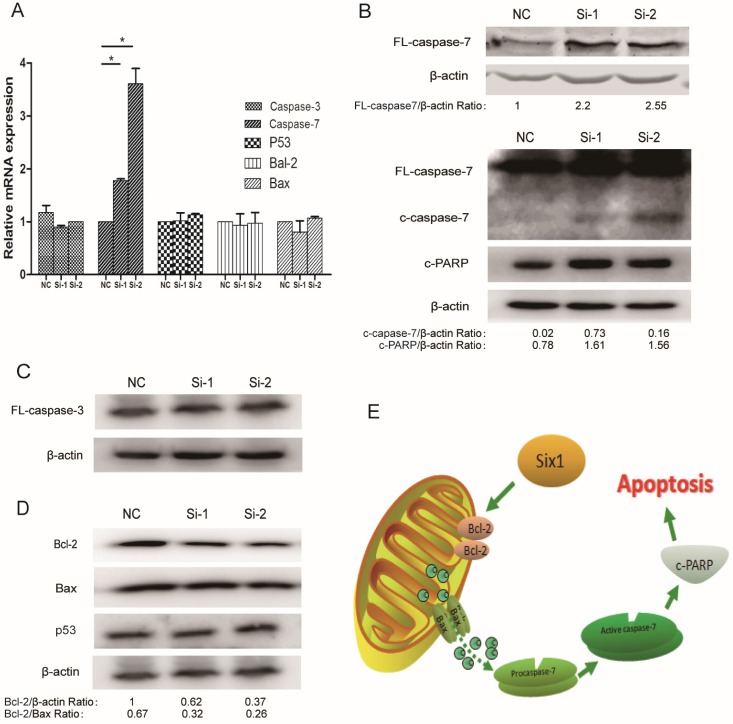
Six1 reduction can inhibit Bcl-2 and activate apoptosis executor caspase-7
To illustrate the apoptosis mechanism, the expressions of some well-established apoptosis related factors were detected when Six1 was silenced. Among these detected factors, both caspase-3 and caspase-7 are considered to mainly execute apoptosis, but our results showed that only caspase-7 mRNA displayed a significant increase in two Six1 knockdown groups (Fig. 4A). Western blot assay confirmed that full-length caspase-7, cleaved caspase-7 and its downstream protein c-PARP were all activated (Fig. 4B), whereas the other important
Figure 4.
Six1 reduction can inhibit Bcl-2 and activate apoptosis executor caspase-7. (A) The relative mRNA levels of apoptosis related factors were detected by qRT-PCR with Six1 silence. (B) Western blot assay detected the protein levels of FL- caspase-7, c-caspase-7 and its downstream effector c-PARP with Six1 reduction. FL and c referred to full length and cleaved proteins respectively. β-actin was the internal control. (C) Caspase-3 protein level was not affected with Six1 knock-down. (D) Bcl-2, Bax, P53 protein expression were detected with Six1 silence. Six1 reduction decreased Bcl-2 protein level but had slight effect on Bax and P53. (E) Schematic diagram displayed the mechanism of Six1-mediated mitochondrial pathway apoptosis.
mitochondrial apoptosis mediator caspase-3 altered slightly (Fig. 4C). That implicated that it was the caspase-7 participating in the Six1-mediated apoptosis process, rather than caspase-3.
Beyond that, we detected the alteration of two crucial proteins in mitochondrial membrane, Bcl-2 and Bax. Bcl-2 and Bax can form heterodimer to regulate Δψm. We found that Six1 reduction was able to down-regulate Bcl-2 protein levels but had no impact on Bax (Fig. 4D). Even so, the decreased ratio of Bcl-2/Bax protein further confirmed that mitochondrial membrane potential have changed toward triggering the apoptosis after Six1 silence, which was consistent with the results of JC-1 test (Fig. 3C to E). As tumor suppressor p53 have been reported to regulate Bcl-2 and Bax expression, herein we also evaluated the alteration of p53 expression. As shown in Fig. 4D, knock-down Six1 did not change p53 expression.
Collectively, as described in Fig. 4E, our findings indicate that Six1 can control Bcl-2/Bax relative expression in mitochondrial membrane, which would alter mitochondrial membrane potential and regulate caspase-7 activation to affect mitochondrial apoptosis during gastric cancer development.
Discussion
Six1 is crucial for normal organogenesis, but also plays vital roles in tumorigenesis. Six1 overexpression can activate proliferation and stimulate transcriptional activation of cyclin A1 in breast tumors directly12, promote tumor lymphangiogenesis by coordinating TGF-β signals to increase VEGF-C expression in cervical cancer 8. In colorectal cancer, epithelial-mesenchymal transition seems to rely on the motivation of ZEB1, which is regulated by Six1 13. Six1 can promote pancreatic cancer by increasing of cyclin D1 14. However, the study of Six1 in gastric cancer is limited. H. Lv et al. have examined Six1 protein expression among 163 gastric adenocarcinomas, 35 gastric dysplasia and 26 normal gastric mucosa with IHC9. They found that positive Six1 protein staining were more frequent in gastric adenocarcinomas and Six1 had the potential to be an independent survival risk factor 9. In this study, we detected Six1 expression by qRT-PCR, Western blot and IHC methods. Our results confirmed that both Six1 transcriptional and protein levels were upregulated in gastric cancer samples (Fig. 1A-D), and Six1 elevation was correlated with lymph node metastasis (p=0.042) and poor differentiation (p=0.039) (Table 1). We also compared Six1 expression in gastritis and different tumor stage patients, and found that Six1 mRNA levels were increased in gastric cancer patients (Fig. 1B). Then, our survival analysis results with online K-M plotter database demonstrated that higher Six1 expression were associated with poor survival outcome significantly (Fig.1E). The similar results were also observed in the lymphatic metastasis and poor differentiation subgroups (Fig.1F, G). Taken together, these findings further establish that Six1 can play vital roles in gastric cancer development, and its functions and biological mechanism need to be elucidated. In the present study, we find Six1 can regulate mitochondrial apoptosis pathway via caspase-7 in gastric cancer.
To date, there are two independent apoptotic signaling cascades: the extrinsic (also named death receptor pathway) and the intrinsic (or called mitochondrial pathway). The extrinsic apoptosis signaling pathways need the involvement of transmembrane receptor-mediated interactions, while the mitochondrial apoptosis pathways are triggered by various of non-receptor-mediated intracellular signals, and mitochondrial apoptosis is the mitochondrial-initiated events15. In mitochondrial pathway, stimuli causes alteration in mitochondrial membrane that causes an opening of the mitochondrial permeability transition (MPT) pore and loss of Δψm. Hypoxia, chemotherapeutic drugs and ionizing radiation mainly trigger a direct activation of the mitochondrial apoptosis pathway. In this study, we found that knocking down Six1 could activate the mitochondrial pathway by JC-1 stain to detect Δψm. During mitochondrial apoptosis pathway, caspase-3 and -7 were testified as two vital executors in response to the loss of Δψm 16. In the present study, we found it was the caspase-7 instead of caspase-3 involved in Six1 regulated apoptosis process in gastric cancer. Our findings are not completely consistent with previous study in osteosarcoma. H. Liu et al. reported that Six1 can promote apoptosis through caspase-3 in osteosarcoma cells 17, but they did not mention the effects of Six1 on caspase-7. The different target genes of Six1 may result from various tissue-specific or developmental stage requirement. Six1 needs caspase-7 to modulate mitochondrial apoptosis in gastric cancer whereas caspase-3 is required for apoptosis in osteosarcoma 17. As for caspase-7, it was not rare for its expression to be regulated independent of caspase-3 actually. For example, R. Hudson et al. reported MiR-106b can down-regulate caspase-7 protein levels in prostate cell lines, while no effect was observed for caspase-318.
Early on, although caspase-7 is widely regarded to act as executioner caspase, its role during the execution phase of apoptosis is obscure, even been questioned 19. However, in recent years, the importance of caspase-7 attracts more attention gradually 16, 18. Biochemical studies discovered that caspase-7 and -3 are able to exhibit various activities toward different protein substrates, but caspase-7 being more selective 19, 20. For example, caspase-7 is more conductive than caspase-3 to the proteolysis of cochaperone p2320. M. Brentnal et al. reported caspase-7 regulates the detachment of cell during mitochondrial apoptosis 21. Studies have demonstrated that caspases family members in stomach cancer are differentially regulated 22. It was reported caspase-3, -8, -9 and -10 are highly expressed, but caspase-2, -6 and -7 are decreased in gastric cancer22. According to our results, the lower expression of caspase-7 may partly attribute to the Six1 upregulation in gastric cancer.
Bcl-2 is an integral membrane protein which mainly locates on the outer membrane of mitochondria. The elevation of Bcl-2 or Bcl-2/Bax ratio are associated with poor long-term outcome 23, 24. Bcl-2 reduction can effectively promote the release of apoptotic effectors from mitochondria and drive apoptosis. In this study, we noticed that the ratio of Bcl-2/Bax was obviously downregulated when Six1 was inhibited. These results further confirmed that Six1 enable to regulate mitochondrial apoptosis pathway. What's more, we found that Six1-induced Bcl-2 alteration was independent of p53 pathway, as Six1 had no effect on p53 expression.
In summary, our results indicate that Six1 is upregulated and associated with poor prognosis and shorter survival time in gastric cancer. Six1 can regulate mitochondrial membrane potential by anti-apoptotic protein Bcl-2, and affect mitochondrial apoptosis via caspase-7. These findings illustrate a novel mechanism of Six1, and suggest that Six1 can serve as a useful prognostic and therapeutic target in gastric cancer.
Acknowledgments
This study was supported by the Natural Science Foundation of China (81071994). Natural Science Foundation of China (81201521, 81672378).
References
- 1.Giordani J, Bajard L, Demignon J, Daubas P, Buckingham M, Maire P. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11310–5. doi: 10.1073/pnas.0611299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–81. doi: 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- 3.Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT. et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8090–5. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SH, Kim J, Ryu JY, Lee S, Yang DK, Jeong D. et al. Transcription coactivator Eya2 is a critical regulator of physiological hypertrophy. Journal of molecular and cellular cardiology. 2012;52:718–26. doi: 10.1016/j.yjmcc.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Tan J, Zhang C, Qian J. Expression and significance of Six1 and Ezrin in cervical cancer tissue. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2011;32:1241–7. doi: 10.1007/s13277-011-0228-8. [DOI] [PubMed] [Google Scholar]
- 6.Ng KT, Lee TK, Cheng Q, Wo JY, Sun CK, Guo DY. et al. Suppression of tumorigenesis and metastasis of hepatocellular carcinoma by shRNA interference targeting on homeoprotein Six1. International journal of cancer Journal international du cancer. 2010;127:859–72. doi: 10.1002/ijc.25105. [DOI] [PubMed] [Google Scholar]
- 7.Sehic D, Karlsson J, Sandstedt B, Gisselsson D. SIX1 protein expression selectively identifies blastemal elements in Wilms tumor. Pediatric blood & cancer. 2012;59:62–8. doi: 10.1002/pbc.24025. [DOI] [PubMed] [Google Scholar]
- 8.Liu D, Li L, Zhang XX, Wan DY, Xi BX, Hu Z. et al. SIX1 promotes tumor lymphangiogenesis by coordinating TGFbeta signals that increase expression of VEGF-C. Cancer research. 2014;74:5597–607. doi: 10.1158/0008-5472.CAN-13-3598. [DOI] [PubMed] [Google Scholar]
- 9.Lv H, Cui A, Sun F, Zhang Y, Li Y, Li L. et al. Sineoculis homeobox homolog 1 protein as an independent biomarker for gastric adenocarcinoma. Experimental and molecular pathology. 2014;97:74–80. doi: 10.1016/j.yexmp.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS one. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coletta RD, Jedlicka P, Gutierrez-Hartmann A, Ford HL. Transcriptional control of the cell cycle in mammary gland development and tumorigenesis. Journal of mammary gland biology and neoplasia. 2004;9:39–53. doi: 10.1023/B:JOMG.0000023587.40966.f6. [DOI] [PubMed] [Google Scholar]
- 13.Ono H, Imoto I, Kozaki K, Tsuda H, Matsui T, Kurasawa Y. et al. SIX1 promotes epithelial-mesenchymal transition in colorectal cancer through ZEB1 activation. Oncogene. 2012;31:4923–34. doi: 10.1038/onc.2011.646. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang L. et al. Six1 promotes proliferation of pancreatic cancer cells via upregulation of cyclin D1 expression. PloS one. 2013;8:e59203. doi: 10.1371/journal.pone.0059203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elmore S. Apoptosis: a review of programmed cell death. Toxicologic pathology. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakhani SA, Masud A, Kuida K, Porter GA Jr, Booth CJ, Mehal WZ. et al. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–51. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua L, Fan L, Aichun W, Yongjin Z, Qingqing C, Xiaojian W. Inhibition of Six1 promotes apoptosis, suppresses proliferation, and migration of osteosarcoma cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:1925–31. doi: 10.1007/s13277-013-1258-1. [DOI] [PubMed] [Google Scholar]
- 18.Hudson RS, Yi M, Esposito D, Glynn SA, Starks AM, Yang Y. et al. MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene. 2013;32:4139–47. doi: 10.1038/onc.2012.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. The Journal of biological chemistry. 2001;276:7320–6. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 20.Walsh JG, Cullen SP, Sheridan C, Luthi AU, Gerner C, Martin SJ. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12815–9. doi: 10.1073/pnas.0707715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brentnall M, Rodriguez-Menocal L, De Guevara RL, Cepero E, Boise LH. Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC cell biology. 2013;14:32. doi: 10.1186/1471-2121-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo NJ, Lee JW, Kim YJ, Soung YH, Kim SY, Nam SW. et al. Loss of caspase-2, -6 and -7 expression in gastric cancers. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2004;112:330–5. doi: 10.1111/j.1600-0463.2004.apm1120602.x. [DOI] [PubMed] [Google Scholar]
- 23.Karakas T, Maurer U, Weidmann E, Miething CC, Hoelzer D, Bergmann L. High expression of bcl-2 mRNA as a determinant of poor prognosis in acute myeloid leukemia. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 1998;9:159–65. doi: 10.1023/a:1008255511404. [DOI] [PubMed] [Google Scholar]
- 24.Bincoletto C, Saad ST, da Silva ES, Queiroz ML. Haematopoietic response and bcl-2 expression in patients with acute myeloid leukaemia. European journal of haematology. 1999;62:38–42. doi: 10.1111/j.1600-0609.1999.tb01112.x. [DOI] [PubMed] [Google Scholar]