Fig. 2.
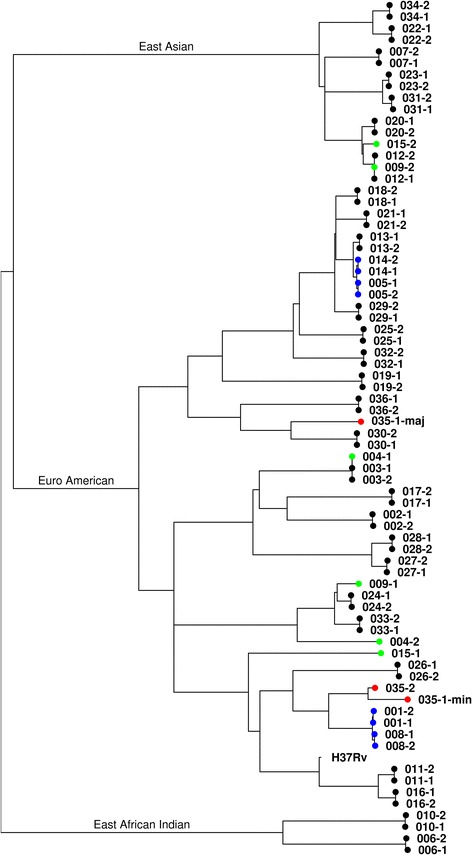
a Phylogenetic reconstruction of 36 pairs of isolates. These were inferred using 5132 high-quality single-nucleotide polymorphisms (SNPs) following the removal of 661,083 low-quality sites and the remaining invariant sites. The tree was rooted using the H37Rv reference strain sequence. Relapse, reinfection and mixed are denoted with black/blue, green and red tips respectively. Blue tip labels are further shown in panels b–e. b–e Branches have been amplified where unexpected similarity was seen; the numbers of SNPs between the most divergent samples are given
