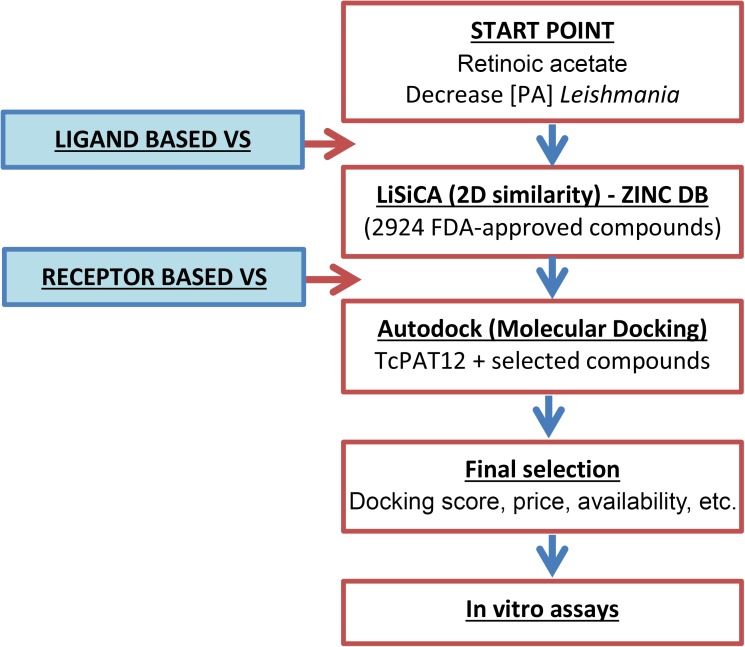
Fig 1. Workflow of the virtual screening strategy.
A two-steps methodology was applied for identification of drug candidates to bind the TcPAT12 substrate recognition site. The ligand-based virtual screening was performed using retinol acetate as reference molecule and a similarity searching algorithm based on the polynomial-time algorithm for finding maximal cliques (LiSiCA) [32]. Tested compounds were obtained from the ZINC database (http://zinc.docking.org/). The second step was a receptor-based strategy using the homology modeled transporter TcPAT12 as the receptor, and selected compounds obtained from the similarity screening as putative ligands. Molecular docking was performed using the AutoDock software [34]. (PA, polyamines; VS, virtual screening).