Figure 3. DXO is a deNADding enzyme in cells and NAD-capped RNAs do not support translation.
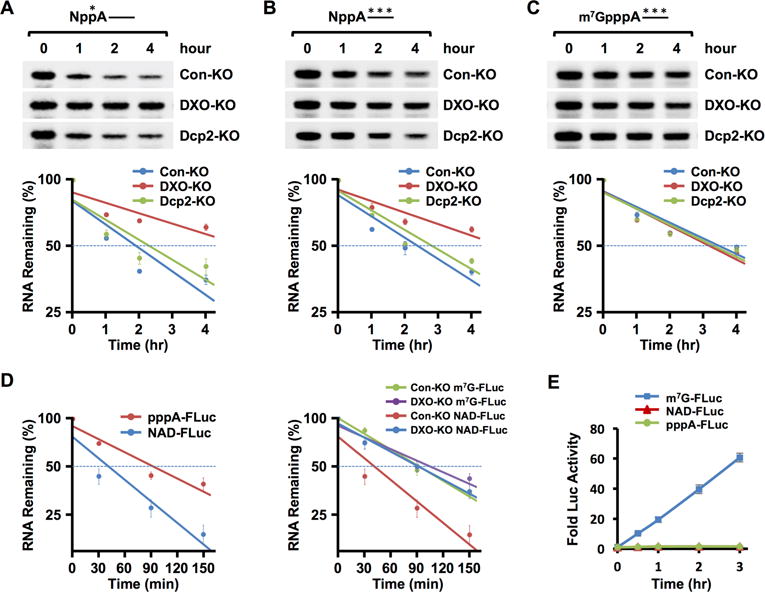
(A) 32P-NAD+ cap-labeled RNA was transfected into HEK293T control knock-out (Con-KO), DXO knock-out (DXO-KO) or Dcp2 knock-out (Dcp2-KO) cell lines with Lipofectamine 3000. Untransfected RNAs were degraded with micrococcal nuclease (MN) and total RNA was isolated at the indicated time points following MN treatment and resolved on 8% Denaturing PAGE and exposed to a PhosphorImager. RNA remaining was quantitated and plotted from three independent experiments with ±SD denoted by the error bars. Stabilities: Con-KO t1/2=2.8 hr [95%CI: 2.0 – 4.5]; DXO-KO t1/2=6.3 hr [95%CI: 4.2 – 11.9] (ANOVA, p=6.10 × 10−4); Dcp2-KO t1/2=3.2 hr, [95%CI: 2.3 – 5.8] (ANOVA, p=0.38). (B) Same as A except the NAD+-capped RNA was uniformly labeled 32P. Stabilities: Con-KO t1/2=3.0 hr [95%CI: 2.4 – 4.3]; DXO-KO t1/2=5.7 hr [95%CI: 4.1 – 8.9] (ANOVA, p=7.88 × 10−4); Dcp2-KO t1/2=3.3 hr, [95%CI: 2.7 – 4.4] (ANOVA, p=0.17). (C) Same as A except the RNA contained an m7G cap and was uniformly labeled within the RNA body with 32P. Stabilities: Con-KO t1/2=4.1 hr [95%CI: 3.2 – 5.8]; DXO-KO t1/2=3.9 hr [95%CI: 3.0 – 5.4] (ANOVA, p=0.51); Dcp2-KO t1/2=4.1 hr, [95%CI: 3.1 – 5.9] (ANOVA, p=0.64). (D). Luciferase RNAs with a 5′-end triphosphate lacking a cap, 5′-end NAD+ cap or m7G cap transfected into control or DXO-KO cells. Cell were treated with micrococcal nuclease to degrade untransfected RNA, harvested at the indicated times and remaining RNA levels quantitated from three independent experiments. The left panel shows RNAs transfections into control HEK293T (Con-KO) cells and the right panel presents RNAs introduced into either Con-KO or DXO-KO cells as indicated. Error bars denoted ±SD. Con-KO m7G-FLuc t1/2= 92.6 min (95% CI: 82.2 to 107.8 min, R2=0.96, p=2.10 × 10−8); DXO-KO m7G-FLuc t1/2=125 min (95% CI: 92.2 to 174 min, R2=0.84, p=1.47 × 10−5); Con-KO NAD-FLuc t1/2= 67.4 min (95% CI: 51.4 to 97.9 min, R2=0.82, p=3.05 × 10−5); DXO-KO NAD-FLuc t1/2= 103.6 min (95% CI: 85.2 to 132.3 min, R2=0.91, p=1.22 × 10−6); Con-KO pppA-RNA=110.7 min (95% CI: 88 to 149.2 min, R2=0.870, p=5.99 × 10−5). (E). Firefly luciferase mRNAs containing either a 5′-end NAD+ cap, m7G cap or no cap and 3′ poly(A)60 tail were co-transfected with m7G-capped Renilla Luciferase RNA into HEK293T cells. Cells were harvested and assayed at the indicated time points. Firefly Luciferase activity was plotted normalized to Renilla Luciferase and data from three independent experiments are presented with error bars representing +/−SD.
