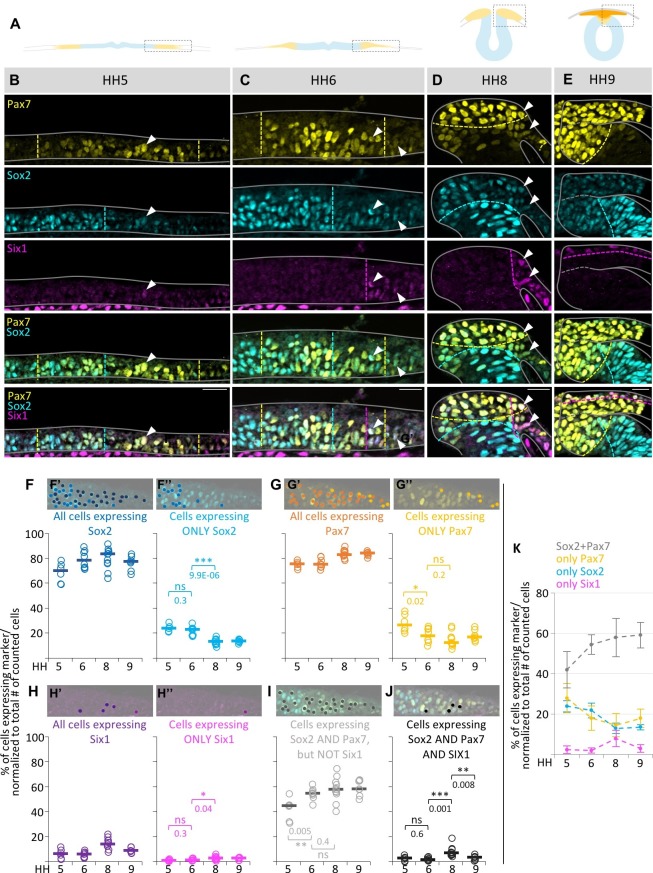
Figure 2. Quantification of marker (co-) expression in single cells.
(A) Schematic diagrams of sections staged accordingly to those used in (B–E). Box indicates area that was imaged in (B–E). (B–E) Transverse sections immunostained for Pax7 (yellow), Sox2 (blue) and Six1 (magenta) at HH5 (B), HH6 (C), HH8 (D) and HH9 (E). Grey lines outline the embryo borders. Dashed colored lines indicate borders between strong and weak/no expression of corresponding markers. Sections are oriented medial (left) to lateral (right). See also Figure 2—figure supplement 1. (F–H) Scatterplots of quantification of all cells that express Sox2 (F’), Pax7 (G’) and Six1 (H’) and the fraction of cells expressing only Sox2 (F’’), Pax7 (G’’) or Six1 (H’’) at HH5 to HH9. (I) Scatterplots representing the fraction of cells that coexpress Sox2 and Pax7, but not Six1 or (J) Sox2, Pax7 and Six1 at HH5 to HH9. See also Figure 2—source data 1. On top of scatterplots are sample images of a HH6 section with dots indicating cells expressing the corresponding marker. (K) Combination of medians of F’’, G’’, H’’ and I to illustrate difference of single (Sox2-blue, Pax7-yellow) or coexpressing cells (Sox2/Pax7-grey) at different stages. Scale bars = 20 µm. Asterisks indicate significance as calculated using a Student’s t-test with p-values displayed. Error bars indicate standard deviation.
DOI: http://dx.doi.org/10.7554/eLife.21620.003
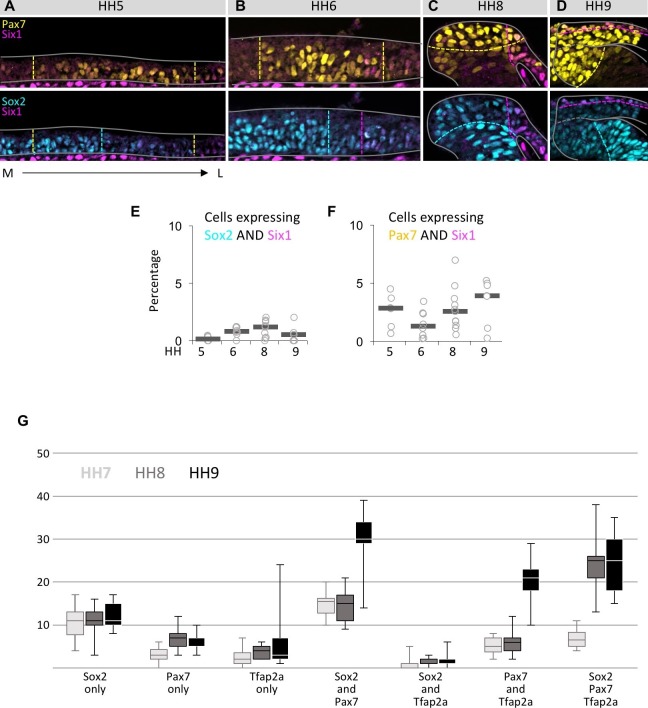
Figure 2—figure supplement 1. Coexpression of markers in single neural plate border cells.
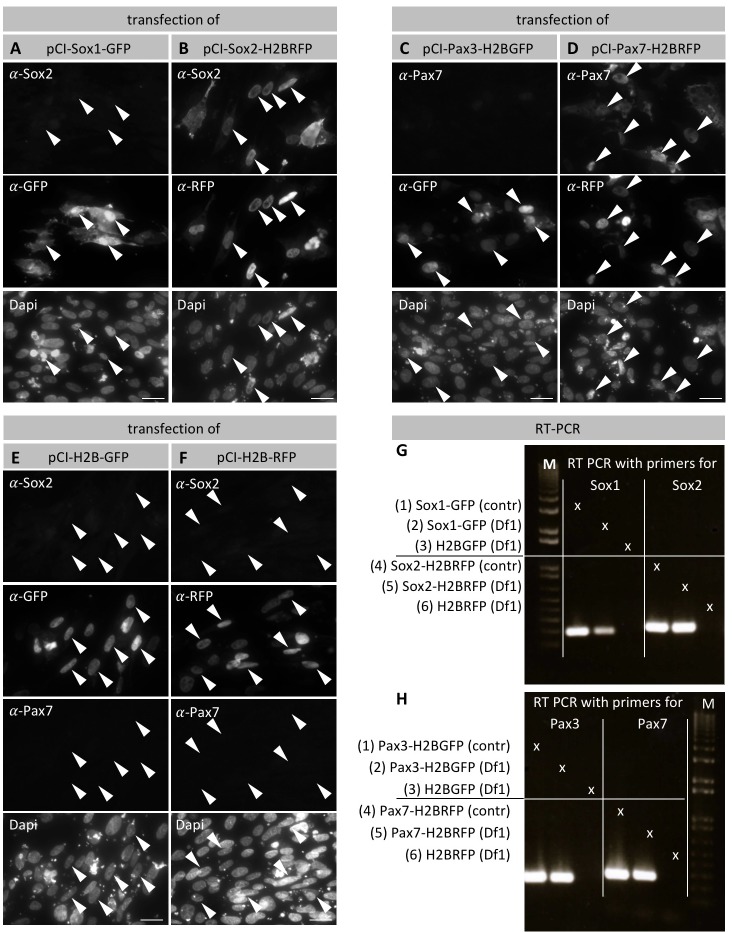
Figure 2—figure supplement 2. Specificity of Sox2 and Pax7 antibodies.
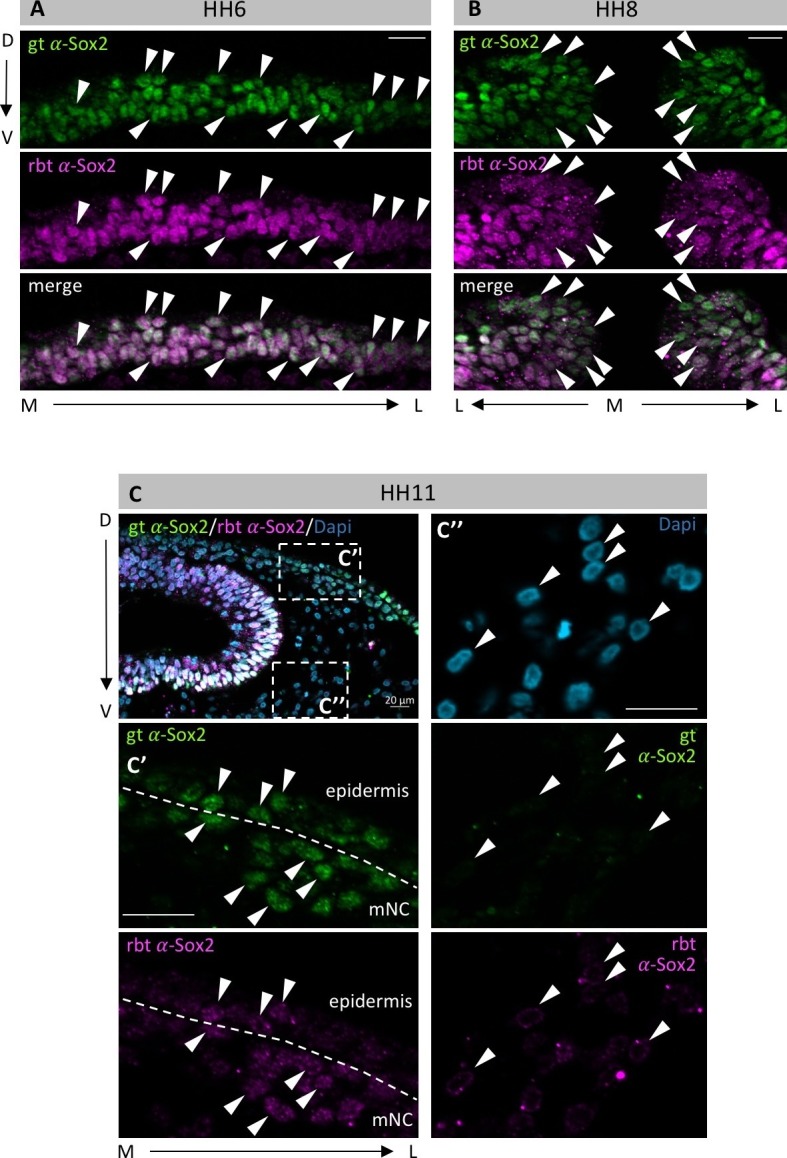
Figure 2—figure supplement 3. Different Sox2 antibodies exhibit identical staining patterns.