Abstract
The Wnt/β-catenin pathway plays a vital role in initiating and sustaining hepatocellular carcinoma (HCC). However, few studies have investigated polymorphisms in the Wnt/β-catenin signaling pathway genes in the Chinese Han population. The aim of the present retrospective study was to investigate the correlations between polymorphisms of the Wnt/β-catenin signaling pathway genes (CTNNB1 and WNT2) and HCC susceptibility, development, and progression.
Twenty tagging single nucleotide polymorphisms were chosen from HapMap data and genotyped in 320 patients with HCC, 320 chronic hepatitis B virus (HBV)-infected patients without HCC (N-HCC, including 95 liver cirrhosis, 164 chronic hepatitis B, and 61 asymptomatic HBV carriers), and 320 healthy controls. Associations between polymorphisms in the 2 Wnt/β-catenin signaling pathway genes (CTNNB1 and WNT2) and HCC susceptibility, development, and progression were investigated.
Genotype AA (P = 0.002, odds ratio [OR] = 2.524) and allele A (P = 0.0003, OR = 1.613) of the WNT2 rs4730775 polymorphism were associated with HCC susceptibility compared with healthy controls. Genotype GA (P = 0.001, OR = 0.567) and allele A (P = 0.002, OR = 0.652) of rs3864004, and genotype AG (P = 0.0004, OR = 0.495) and allele G (P = 0.001, OR = 0.596) of rs11564475 in the CTNNB1 gene were correlated with HCC compared with N-HCC patients. These findings were consistent in dominant and recessive models. Multidimensionality reduction analysis revealed that interactions among rs3864004, rs11564475, and rs4730775 were significantly associated with HCC compared with N-HCC patients. The polymorphism rs4135385 of CTNNB1 genotype GA was associated with a higher risk for Stage III + IV HCC (modified Union for International Cancer Control) (P = 0.001, OR = 2.238).
Genetic polymorphisms in the WNT2 and CTNNB1 genes were closely associated with HCC risk and progression in a Chinese Han population.
Keywords: CTNNB1, genetic polymorphism, hepatocellular carcinoma, WNT2
1. Introduction
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related deaths worldwide, accounting for 70% to 85% of all liver cancers. About 75% to 80% of HCC cases are caused by chronic hepatitis B virus (HBV) or hepatitis C virus infection.[1] China is an endemic region for chronic HBV infection with 94 million patients, and approximately half of the global HCC patients are in China.[2,3] Overall, 2% to 3% of chronic HBV-infected patients develop HCC.[4] Male sex,[5,6] old age, inflammation,[7] and cirrhosis are known risk factors for HCC.[5,8,9] Moreover, single nucleotide polymorphisms (SNPs) of host genes have been found to be correlated with the development and progression of HCC.[10–12]
The Wnt/β-catenin pathway plays a vital role in initiating and sustaining HCC. It plays an important role in embryogenesis and in adult tissue homeostasis.[13] The CTNNB1 gene encodes β-catenin protein, which plays an important role in tissue regeneration, proliferation, differentiation, and development.[14,15] Wnt2 is a member of the Wnt/β-catenin pathway, and has been reported to be associated with multiple cancers.[16–19] Although the Wnt/β-catenin signaling pathway is important for HCC, few studies have investigated polymorphisms in the Wnt/β-catenin signaling pathway genes in the Chinese Han population.
The aim of the current retrospective study was to investigate the correlations between variants in 2 Wnt/β-catenin signaling pathway genes (CTNNB1 and WNT2) and HCC susceptibility and progression. SNP–SNP interactions were analyzed by multidimensionality reduction (MDR).
2. Methods
2.1. Patients
In the present study, we obtained blood samples from unrelated Chinese Han patients at the Liaocheng People's Hospital, Liaocheng, Shandong Province, China between January 2012 and December 2015. A total of 960 individuals were selected for the study, including 320 chronic HBV-infected patients with HCC, 320 chronic HBV-infected patients without HCC (N-HCC, including 95 liver cirrhosis, 164 chronic hepatitis B [CHB], and 61 asymptomatic HBV carriers), and 320 healthy controls (HC). All the HCC and N-HCC cases showed more than 6 months of sero-positivitiy for hepatitis B surface antigen. We diagnosed HCC and liver cirrhosis patients according to clinical and biological criteria, which were confirmed by iconography examination (magnetic resonance imaging or computed tomography). Asymptomatic HBV carriers were defined as patients with chronic HBV infection and persistently normal liver biochemical parameters. CHB patients were defined as chronic HBV-infected patients with abnormal liver biochemical parameters (intermittent or persistent elevations of bilirubin or aminotransferases), and no evidence of HCC or liver cirrhosis. The HC group was defined as individuals with negative HBV sero-markers, no history of hepatitis, normal liver function, and no cancers or other disorders.
Each HC individual was matched with an HCC patient and an N-HCC patient by sex. The study protocol was approved by the ethics committee of Liaocheng People's Hospital. Written informed consent was obtained from all subjects.
2.2. Tagging SNP selection
Twenty tagging SNPs (tgSNPs) were chose from the CTNNB1 and WNT2 genes among data in the International HapMap Project, release27 (http://hapmap.ncbi.nlm.nih.gov) for the CHB population. The exclusion criteria were an r2 value < 0.8 and a minimum allele frequency <0.05. Nonsynonymous SNPs or functional SNPs in the literature or in the National Center for Biotechnology Information database were selected initially. The F-SNP program was used to determine the functional SNPs (http://compbio.cs.queensu.ca/F-SNP/).
2.3. DNA purification and SNP analysis
Wizard Genomic DNA Purification Kit (Promega; Madison, WI, USA) was used to obtain genomic DNA samples from peripheral blood leukocytes, according to the manufacturer's instructions. A NanoDrop spectrophotometer was used to measure the DNA concentration. DNA samples were diluted to 50 ng/μL and stored at −80°C for genotyping analysis. We used IplexGOLD chemistry on a SEQUENOM mass spectrometer (Sequenom Inc, San Diego, CA) to genotype the selected 20 tgSNPs. A negative control and duplicate samples were set in every 96-well plate for quality control.
2.4. SNP–SNP interaction analysis
Potential interactions among SNPs were tested by MDR (version 3.0.2), which is a nonparametric program. The program contains permutation testing and cross-validation. MDRpt (version 1.0_beta_2) was used for permutation testing to determine statistical significance. Significant interactions were defined according to P < 0.05. The best model is one that shows a cross-validation consistency (CVC) ≥ 9 and the largest testing balance accuracy (TBA). The MDRpt and MDR programs are freely available from http://www.epistasis.org.
2.5. Statistical analysis
HCC risk was investigated according to the genotypes and alleles of CTNNB1 and WNT2 in comparison to the control groups. We tested for Hardy–Weinberg equilibrium in the HC group with IHG program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). The Mann–Whitney U test or χ2 test was used to test differences in age, sex, or clinical characteristics between groups. We used recessive or dominant genetic models to evaluate the associations of each genotype with HCC. The relative risks of SNPs for HCC were investigated with binary logistic regression. Haploview software (v4.2) was used to identify haplotype frequencies. Bonferroni correction was used to correct P values for multiple testing (0.05/20 = 0.0025). Statistical calculations were carried out by SPSS 21.0 (Chicago, IL), and P < 0.0025 was considered to indicate statistical significance.
3. Results
Two SNPs (rs2293302 from CTNNB1 and rs733153 from WNT2) were excluded from the analysis owing to genotyping success rates <92%. The other 18 SNPs selected were all in accordance with Hardy–Weinberg equilibrium in the HC group.
3.1. Characteristics of the study population
Characteristics of the study population among the HCC, N-HCC, and HC groups are shown in Table 1. Sex and age did not differ significantly among the 3 groups (P > 0.05). The HCC group had significantly higher levels of HBV DNA, total bilirubin (TBIL), aspartate aminotransferase (AST), and alanine aminotransferase (ALT) compared with the HC group. However, the levels of HBV DNA, TBIL, AST, and ALT did not differ significantly between the N-HCC and HCC groups.
Table 1.
Demographic and clinical characteristics of the study population.

3.2. Correlations of SNPs with HCC
Compared with the HC group, the rs4730775 genotype AA (P = 0.002, odds ratio [OR] = 2.524) and allele A (P = 0.0003, OR = 1.613) from the WNT2 gene showed significant differences in the HCC group. Significant associations of this SNP with HCC susceptibility were also detected in the dominant and recessive genetic models (P = 0.005, OR = 1.576; P = 0.005, OR = 2.272) (Table 2).
Table 2.
Association of WNT2 (rs4730775) in HCC risk.
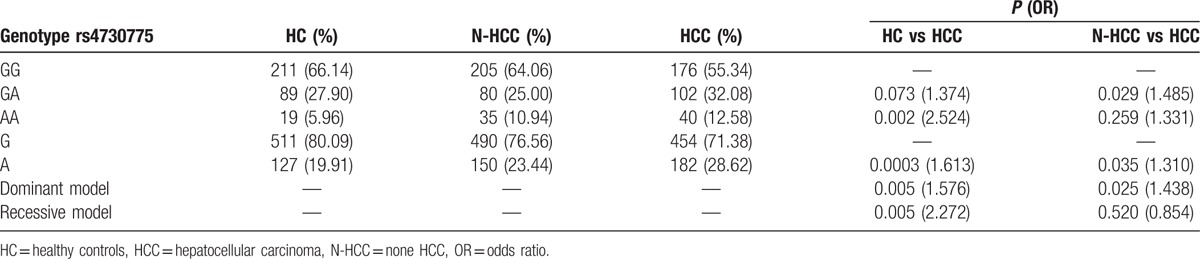
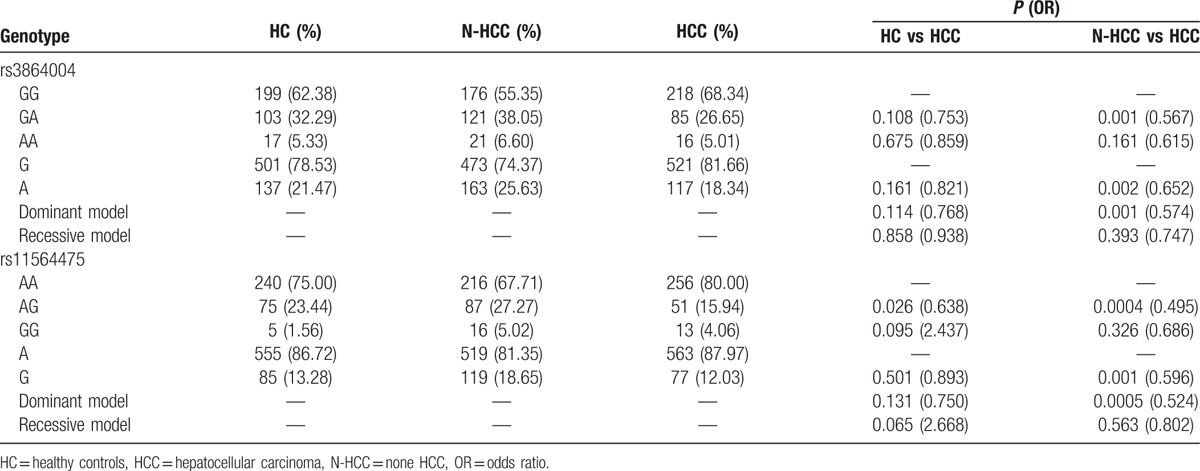
Compared with the N-HCC group, the rs3864004 genotype GA (P = 0.001, OR = 0.567) and allele A (P = 0.002, OR = 0.652) and the rs11564475 genotype AG (P = 0.0004, OR = 0.495) and allele G (P = 0.001, OR = 0.596) of the CTNNB1 gene showed significant differences in the HCC group. Significant associations of these SNPs with HCC were also detected in the dominant and recessive genetic models (P = 0.001, OR = 0.574; P = 0.0005, OR = 0.524) (Table 3).
Table 3.
Association of CTNNB1 (rs3864004 and rs11564475) in HCC risk.
3.3. Correlations of SNPs with HCC tumor stage
Genotype GA of the rs4135385 polymorphism of CTNNB1 was associated with a higher risk for Stage III + IV HCC (modified Union for International Cancer Control) (P = 0.001, OR = 2.238). This finding was confirmed in the dominant model (P = 0.001, OR = 2.207). We did not find any other SNPs associated with HCC tumor stage (Table 4).
Table 4.
Association of CTNNB1 rs4135385 in HCC tumor stage.

3.4. MDR for SNP–SNP interactions
MDR was performed to analyze the SNP–SNP interactions. Since rs4730775, rs3864004, and rs11564475 were associated with HCC, we carried out MDR for these 3 SNPs with HCC as the case group and HC or N-HCC as the control group. In this analysis, we found 1-way (rs3864004), 2-way (rs11564475 and rs4730775), and 3-way (rs3864004, rs11564475, and rs4730775) interactions in the HCC versus N-HCC comparison (Table 5). The best model was that for the 3-way interaction among rs3864004, rs11564475, and rs4730775 (TBA of 0.5938 and CVC 10/10; P < 0.001) (Fig. 1).
Table 5.
MDR interaction analysis between SNPs in N-HCC versus HCC.
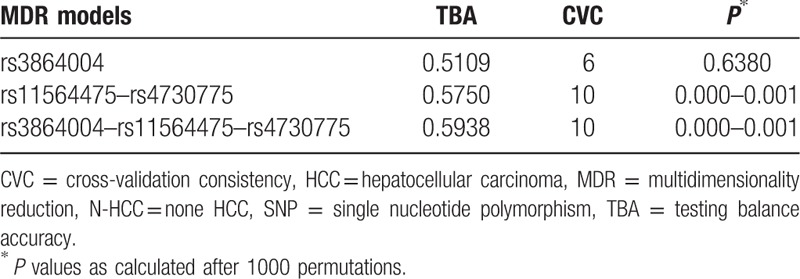
Figure 1.
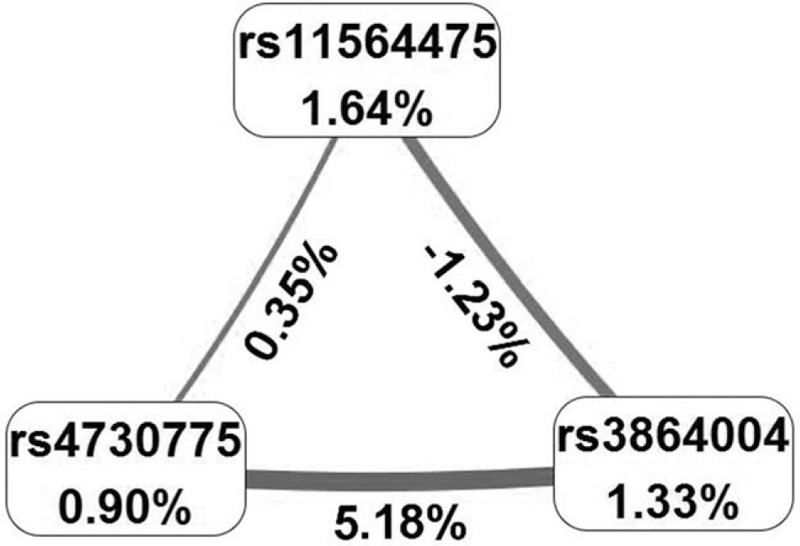
Interactions among SNPs in relation to HCC development. Interactions among SNPs in HCC versus N-HCC. Values in nodes represent the main effect. Values between nodes represent the interaction effect. HCC = hepatocellular carcinoma, N-HCC = chronic HBV-infected patients without HCC, SNP = single nucleotide polymorphism.
4. Discussion
In the present study, 20 tgSNPs from the WNT2 and CTNNB1 genes were genotyped in a Chinese Han population. One SNP of the WNT2 gene (rs4730775) and 2 SNPs of the CTNNB1 gene (rs11564475 and rs3864004) were found to be correlated with HCC. Moreover, rs4135385 of the CTNNB1 gene was associated with HCC tumor stage. Interactions among the 3 SNPs (rs3864004, rs11564475, and rs4730775) showed an association with HCC risk.
Accumulating evidence has revealed that Wnt/β-catenin signaling plays an important role in hepatic carcinogenesis.[20] Polymorphisms of CTNNB1 were reported to be associated with prostate cancer,[21] breast cancer,[22] colorectal cancer,[23] and gastric cancer.[24] In a Korean population, CTNNB1 polymorphisms were suggested to be involved in tumor development and survival of HBV-associated HCC patients.[25] However, there are no previous studies related to polymorphisms in CTNNB1 with HBV-related HCC patients in the Chinese Han population. In the present study, we found that genetic polymorphisms of CTNNB1 (rs11564475 and rs3864004) were associated with HCC development. rs11564475 is located in the intron region (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=11564475) and rs3864004 is located in the promoter region (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ ref.cgi?rs=3864004) of CTNNB1. The functional prediction scores (F-SNP) for the 2 SNPs were 0.268 (rs11564475) and 0.065 (rs3864004). rs3864004 was reported to affect CTNNB1 expression levels in Korean asthma patients.[26] We speculate that both rs11564475 and rs386400 have functional effects and could influence the CTNNB1 expression level. However, direct experiments are needed to validate the functional consequences of these SNPs.
Although WNT2 is reported to be associated with multiple cancers, few studies have investigated polymorphisms of WNT2 with HBV-related HCC. In the present study, a polymorphism of WNT2 (rs4730775) was found to be correlated with HCC susceptibility. This is the first study to reveal that rs4730775 is associated with HBV-related HCC. rs4730775 is located in the 3′-untranslated region (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=4730775) of WNT2. The functional prediction score (F-SNP) for rs4730775 was 0.5. We speculate that rs4730775 might have functional consequences and could affect the WNT2 expression level. However, direct experiments are needed to validate its function.
MDR analysis revealed that rs3864004 (CTNNB1), rs11564475 (CTNNB1), and rs4730775 (WNT2) might interact in HBV-related HCC development. Both gene products (CTNNB1 and WNT2) are involved in the Wnt/β-catenin signaling pathway. Therefore, all 3 SNPs might affect the expression of CTNNB1 and WNT2, resulting in HBV-related HCC development.
A previous study showed that mutation in the CTNNB1 gene could affect ß-catenin activity, which is correlated with liver tumor progression.[27] This is consistent with our finding that rs4135385 of the CTNNB1 gene was associated with HCC tumor stage. rs4135385 is located in the intron of CTNNB1 (http://www.ncbi.nlm.nih.gov/proje cts/SNP/snp_ref.cgi?rs=4135385), and the functional prediction score (F-SNP) for rs4135385 was 0.242. Introns play a vital role in the regulation of transcription or post-transcription processes to affect gene expression.[28,29] Therefore, SNPs in the intron region could affect this regulation. We speculate that rs4135385 might be a functional SNP to affect the expression of CTNNB1.
4.1. Limitations
There are 3 limitations of this study that should be taken into consideration. First, all included individuals (HC, N-HCC, and HCC patients) were recruited from only 1 hospital, which might result in selection bias. However, the distributions of all genotypes were in accordance with Hardy–Weinberg equilibrium in the HC group, except for 2 SNPs (rs2293302 from CTNNB1 and rs733153 from WNT2), which revealed that our subjects might be a good representation of the general population. Second, our study had a relatively small sample size, which might reduce the ability to find differences among groups in statistical analysis. Third, our study was a retrospective study, which had a less power to assess the risk of HCC development than a prospective study, in particular in cirrhotic HBV-infected patients (at high risk for liver cancer).
5. Conclusion
In conclusion, we found that polymorphisms in the Wnt/β-catenin signaling pathway genes (CTNNB1 and WNT2) are associated with HCC susceptibility and progression. In the future, more studies (especially prospective study) in multiple centers with larger sample sizes are needed to validate our findings that genetic polymorphisms in WNT2 and CTNNB1 are closely associated with HCC risk.
Footnotes
Abbreviations: ALT = alanine aminotransferase, AST = aspartate aminotransferase, CHB = chronic hepatitis B, CVC = cross-validation consistency, HBV = hepatitis B virus, HC = healthy controls, HCC = hepatocellular carcinoma, MDR = multidimensionality reduction, N-HCC = chronic HBV-infected patients without HCC, OR = odds ratio, SNP = single nucleotide polymorphism, TBA = Testing balance accuracy, TBIL = total bilirubin, TgSNP = tagging SNP.
The authors have no funding and conflicts of interest to disclose.
References
- [1].Lok AS. Does antiviral therapy for hepatitis B and C prevent hepatocellular carcinoma? J Gastroenterol Hpatol 2011;26:221–7. [DOI] [PubMed] [Google Scholar]
- [2].Yin J, Zhang H, He Y, et al. Distribution and hepatocellular carcinoma-related viral properties of hepatitis B virus genotypes in Mainland China: a community-based study. Cancer Epidemiol Biomarkers Prevent 2010;19:777–86. [DOI] [PubMed] [Google Scholar]
- [3].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [4].El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365:1118–27. [DOI] [PubMed] [Google Scholar]
- [5].Bolondi L, Sofia S, Siringo S, et al. Surveillance programme of cirrhotic patients for early diagnosis and treatment of hepatocellular carcinoma: a cost effectiveness analysis. Gut 2001;48:251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zaman SN, Melia WM, Johnson RD, et al. Risk factors in development of hepatocellular carcinoma in cirrhosis: prospective study of 613 patients. Lancet (London, England) 1985;1:1357–60. [DOI] [PubMed] [Google Scholar]
- [7].Tarao K, Rino Y, Ohkawa S, et al. Association between high serum alanine aminotransferase levels and more rapid development and higher rate of incidence of hepatocellular carcinoma in patients with hepatitis C virus-associated cirrhosis. Cancer 1999;86:589–95. [DOI] [PubMed] [Google Scholar]
- [8].Degos F, Christidis C, Ganne-Carrie N, et al. Hepatitis C virus related cirrhosis: time to occurrence of hepatocellular carcinoma and death. Gut 2000;47:131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tsai JF, Jeng JE, Ho MS, et al. Effect of hepatitis C and B virus infection on risk of hepatocellular carcinoma: a prospective study. Br J Cancer 1997;76:968–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Qu L-S, Jin F, Guo Y-M, et al. Nine susceptibility loci for hepatitis B virus-related hepatocellular carcinoma identified by a pilot two-stage genome-wide association study. Oncol Lett 2016;11:624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jiang D-K, Sun J, Cao G, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet 2013;45:72–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Peng H, Li QL, Hou SH, et al. Association of genetic polymorphisms in CD8+ T cell inhibitory genes and susceptibility to and progression of chronic HBV infection. Infect Genet Evolut 2015;36:467–74. [DOI] [PubMed] [Google Scholar]
- [13].Takigawa Y, Brown AM. Wnt signaling in liver cancer. Curr Drug Targets 2008;9:1013–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol 1998;14:59–88. [DOI] [PubMed] [Google Scholar]
- [15].Moon RT, Bowerman B, Boutros M, et al. The promise and perils of Wnt signaling through beta-catenin. Science 2002;296:1644–6. [DOI] [PubMed] [Google Scholar]
- [16].Park JK, Song JH, He TC, et al. Overexpression of Wnt-2 in colorectal cancers. Neoplasma 2009;56:119–23. [DOI] [PubMed] [Google Scholar]
- [17].Cheng XX, Wang ZC, Chen XY, et al. Correlation of Wnt-2 expression and beta-catenin intracellular accumulation in Chinese gastric cancers: relevance with tumour dissemination. Cancer Lett 2005;223:339–47. [DOI] [PubMed] [Google Scholar]
- [18].Wang W, Xue L, Liu H, et al. Aberrant changes of Wnt2/beta-catenin signaling pathway induced by sodium nitroprusside in human esophageal squamous cell carcinoma cell lines. Cancer Invest 2010;28:230–41. [DOI] [PubMed] [Google Scholar]
- [19].Mazieres J, You L, He B, et al. Wnt2 as a new therapeutic target in malignant pleural mesothelioma. Int J Cancer 2005;117:326–32. [DOI] [PubMed] [Google Scholar]
- [20].Bengochea A, de Souza MM, Lefrancois L, et al. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br J Cancer 2008;99:143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang SP, Ting WC, Chen LM, et al. Association analysis of Wnt pathway genes on prostate-specific antigen recurrence after radical prostatectomy. Ann Surg Oncol 2010;17:312–22. [DOI] [PubMed] [Google Scholar]
- [22].Jia YM, Xie YT, Wang YJ, et al. Association of genetic polymorphisms in CDH1 and CTNNB1 with breast cancer susceptibility and patients’ prognosis among Chinese Han women. PLoS ONE 2015;10:e0135865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Starinsky S, Figer A, Ben-Asher E, et al. Genotype phenotype correlations in Israeli colorectal cancer patients. Int J Cancer 2005;114:58–73. [DOI] [PubMed] [Google Scholar]
- [24].Wang S, Tian Y, Wu D, et al. Genetic variation of CTNNB1 gene is associated with susceptibility and prognosis of gastric cancer in a Chinese population. Mutagenesis 2012;27:623–30. [DOI] [PubMed] [Google Scholar]
- [25].Kim SS, Cho HJ, Lee HY, et al. Genetic polymorphisms in the Wnt/beta-catenin pathway genes as predictors of tumor development and survival in patients with hepatitis B virus-associated hepatocellular carcinoma. Clin Biochem 2016;49:792–801. [DOI] [PubMed] [Google Scholar]
- [26].Bae S, Lee H, Choi BW, et al. Beta-catenin promoter polymorphism is associated with asthma risk in Korean subjects. Clin Biochem 2012;45:1187–91. [DOI] [PubMed] [Google Scholar]
- [27].Rebouissou S, Franconi A, Calderaro J, et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different ss-catenin activity associated with liver tumor progression. Hepatology (Baltimore, MD) 2016;64:2047–61. [DOI] [PubMed] [Google Scholar]
- [28].Miller AD, Bender MA, Harris EA, et al. Design of retrovirus vectors for transfer and expression of the human beta-globin gene. J Virol 1988;62:4337–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ismail SI, Kingsman SM, Kingsman AJ, et al. Split-intron retroviral vectors: enhanced expression with improved safety. J Virol 2000;74:2365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]


