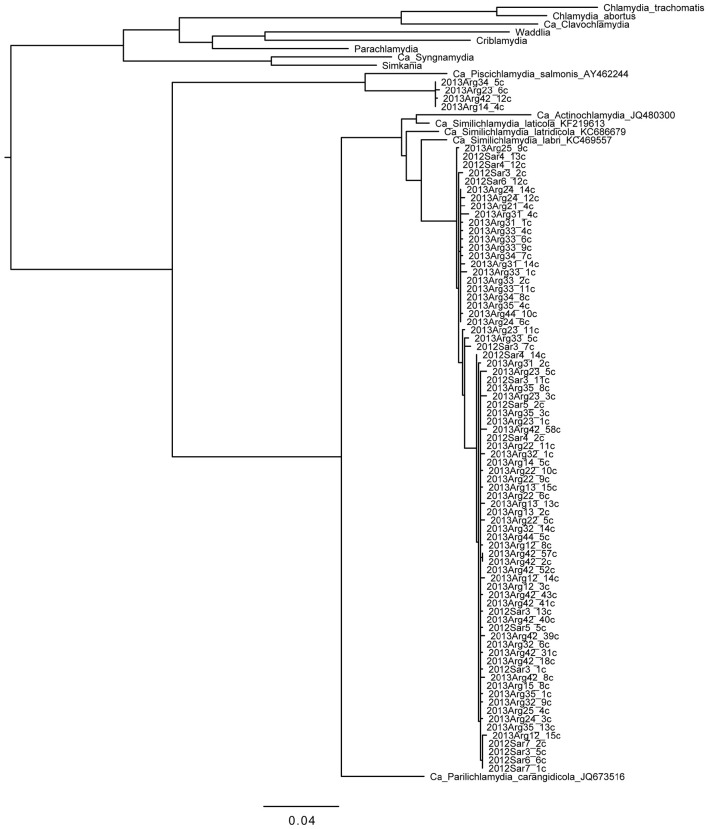
Figure 1.
Phylogenetic tree of chlamydial 16S rRNA gene sequences obtained from gilthead seabream gills. Reference sequences from the phylum as a whole and from the deep-rooted “fish-Chlamydia” clade (provisional families Ca. Piscichlamydiae, Parilichlamydiae, Actinochlamydiae, and Similichlamydiae) are shown to put the sequences we obtained in this study in context. Clones from fish samples are named from the gill sample with the postscript “c” denoting amplification with Chlamydiae-specific primers. Four of the novel sequences from four fish from Argolida are more closely related to Ca. P. salmonis; 63 of the novel sequences from Saronikos and Argolida are more closely related to Ca. S. labri and Ca. S. latridicola. Sequences (~1,100 bp) were aligned and phylogenies generated within Seaview with 100 bootstraps. Branches separating the novel sequences from the reference sequences have bootstraps of 97% (Piscichlamydiaceae clade) and 100% (Similichlamydiaceae clade). Scale bar indicates number of substitutions per site.