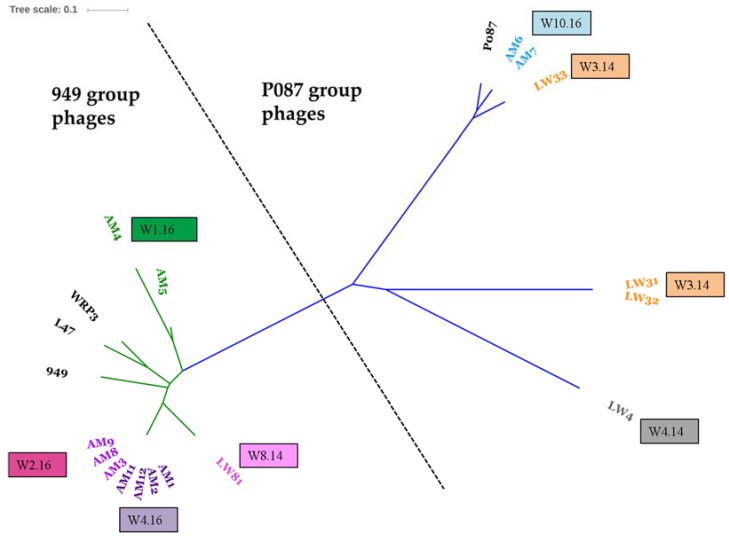
Figure 1.
Unrooted phylogenetic tree of the complete nucleotide sequences of the 949 (green branches) and P087 (blue branches) isolates from this study and previously sequenced members 949, L47, WRP3, and P087. Whole phage nucleotide alignment was performed via ClustalW V2.1. Subsequently, the phylogenetic tree was generated using the neighbor-joining method and bootstrapped (×1000) replicates. Visualisation of the phylogenetic tree was performed using the Itol software (http://itol.embl.de/). Clusters of phage genomes by source sample are indicated in coloured text and corresponding coloured text boxes with the sample from which the phages were derived. The 949 phages isolated from W1.16 (green) are genetically distinct from those isolated from samples W2.16 (hot pink), W4.16 (purple) and W8.14 (pale pink). Similarly, the P087 phage isolates are genetically distinct based on the sample source with the exception of LW33 (isolated from W3.14, orange), which groups more closely to the 2016 isolates from W10.16 (blue) and P087, while phage LW4 occupies a distinct position (grey) highlighting its more distant relationship to the other P087 isolates.